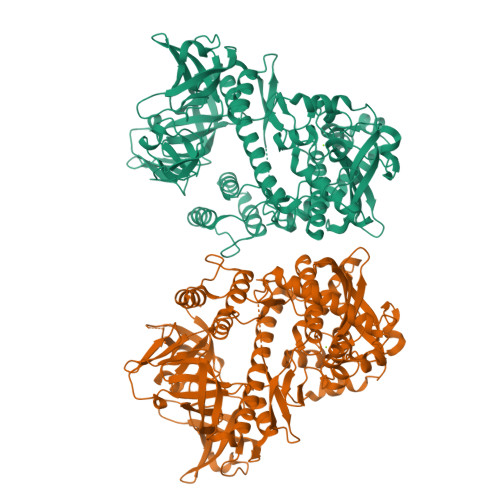
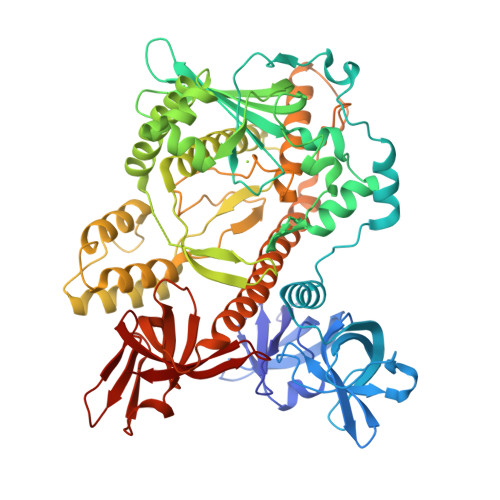
Structural insights into RNA unwinding and degradation by RNase R.
Chu, L.Y., Hsieh, T.J., Golzarroshan, B., Chen, Y.P., Agrawal, S., Yuan, H.S.(2017) Nucleic Acids Res 45: 12015-12024
- PubMed: 29036353
- DOI: https://doi.org/10.1093/nar/gkx880
- Primary Citation of Related Structures:
5XGU - PubMed Abstract:
RNase R is a conserved exoribonuclease in the RNase II family that primarily participates in RNA decay in all kingdoms of life. RNase R degrades duplex RNA with a 3' overhang, suggesting that it has RNA unwinding activity in addition to its 3'-to-5' exoribonuclease activity. However, how RNase R coordinates RNA binding with unwinding to degrade RNA remains elusive. Here, we report the crystal structure of a truncated form of Escherichia coli RNase R (residues 87-725) at a resolution of 1.85 Å. Structural comparisons with other RNase II family proteins reveal two open RNA-binding channels in RNase R and suggest a tri-helix 'wedge' region in the RNB domain that may induce RNA unwinding. We constructed two tri-helix wedge mutants and they indeed lost their RNA unwinding but not RNA binding or degrading activities. Our results suggest that the duplex RNA with an overhang is bound in the two RNA-binding channels in RNase R. The 3' overhang is threaded into the active site and the duplex RNA is unwound upon reaching the wedge region during RNA degradation. Thus, RNase R is a proficient enzyme, capable of concurrently binding, unwinding and degrading structured RNA in a highly processive manner during RNA decay.
Organizational Affiliation:
Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan 11529, ROC.