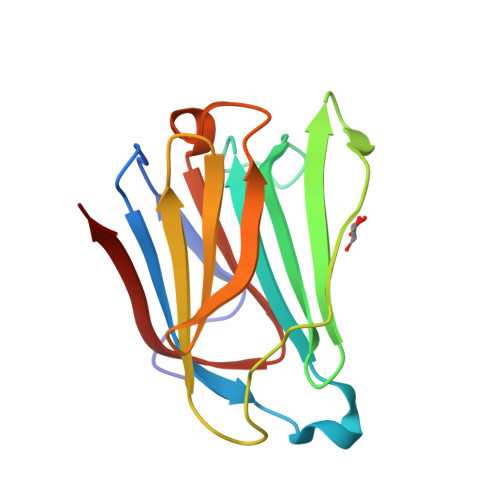
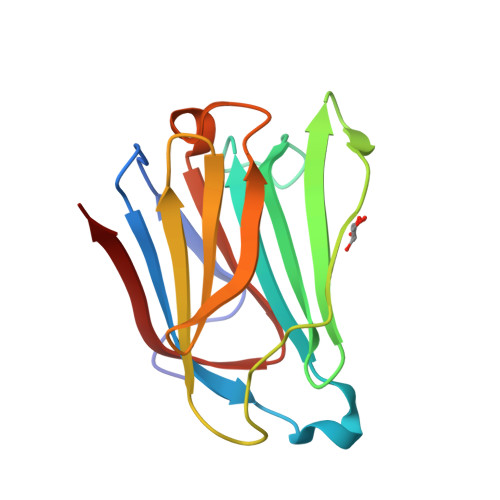
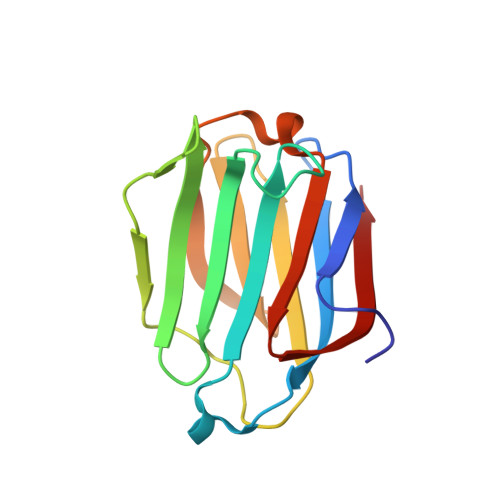
Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Su, J., Wang, Y., Si, Y., Gao, J., Song, C., Cui, L., Wu, R., Tai, G., Zhou, Y.(2018) Sci Rep 8: 980-980
- PubMed: 29343868
- DOI: https://doi.org/10.1038/s41598-018-19465-0
- Primary Citation of Related Structures:
5XG7, 5XG8, 5Y03 - PubMed Abstract:
During pregnancy, placental protein-13 (galectin-13) is highly expressed in the placenta and fetal tissue, and less so in maternal serum that is related to pre-eclampsia. To understand galectin-13 function at the molecular level, we solved its crystal structure and discovered that its dimer is stabilized by two disulfide bridges between Cys136 and Cys138 and six hydrogen bonds involving Val135, Val137, and Gln139. Native PAGE and gel filtration demonstrate that this is not a crystallization artifact because dimers also form in solution. Our biochemical studies indicate that galectin-13 ligand binding specificity is different from that of other galectins in that it does not bind β-galactosides. This is partly explained by the presence of Arg53 rather than His53 at the bottom of the carbohydrate binding site in a position that is crucial for interactions with β-galactosides. Mutating Arg53 to histidine does not re-establish normal β-galactoside binding, but rather traps cryoprotectant glycerol molecules within the ligand binding site in crystals of the R53H mutant. Moreover, unlike most other galectins, we also found that GFP-tagged galectin-13 is localized within the nucleus of HeLa and 293 T cells. Overall, galectin-13 appears to be a new type of prototype galectin with distinct properties.
Organizational Affiliation:
Jilin Province Key Laboratory for Chemistry and Biology of Natural Drugs in Changbai Mountain, School of Life Sciences, Northeast Normal University, Changchun, 130024, PR China.