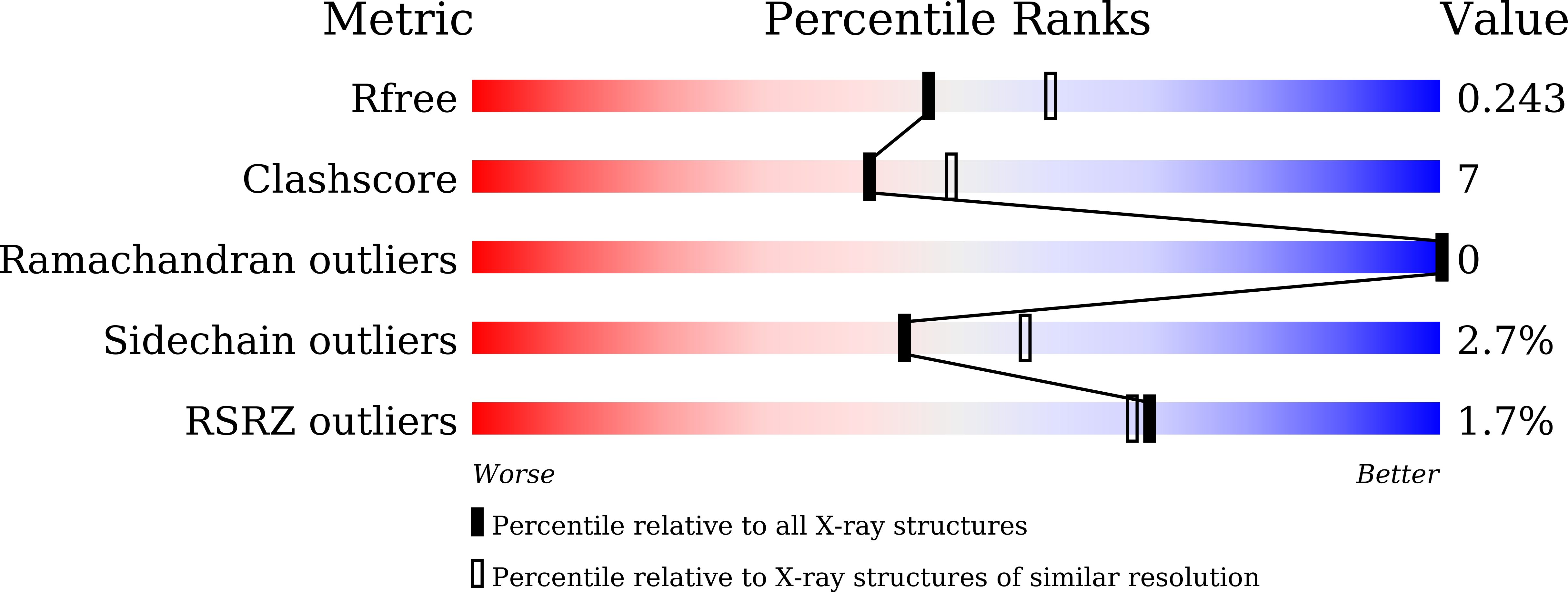
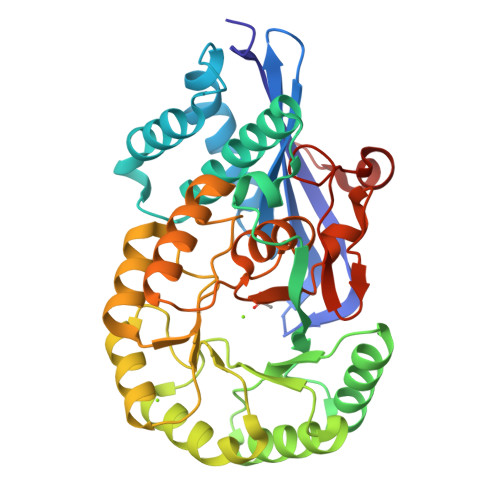
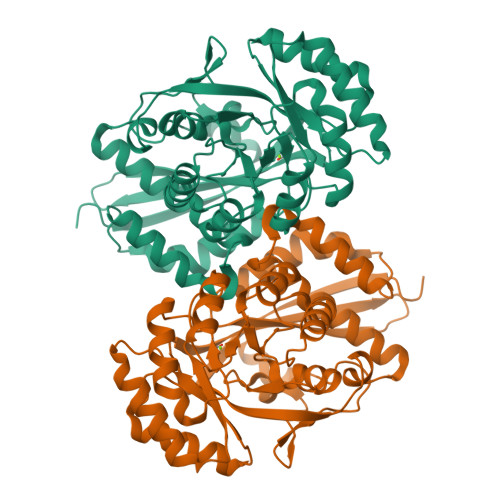
Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily.
Lee, S., Kim, K.H., Kim, H.-Y., Choi, I.-G.(2017) Biochem Biophys Res Commun 491: 217-222
- PubMed: 28716734
- DOI: https://doi.org/10.1016/j.bbrc.2017.07.080
- Primary Citation of Related Structures:
5XD7, 5XD8, 5XD9 - PubMed Abstract:
3,6-Anydro-l-galatonate cycloisomerase (ACI) catalyzes the cycloisomerization of a 3,6-anhydro-l-galactonic acid known as a novel metabolite in agarolytic bacteria. Here, we present 3-D structures of ACI from Vibrio sp. strain EJY3 (VejACI) in native and mutant forms at 2.2 Å and 2.6 Å resolutions, respectively. The enzyme belongs to the mandelate racemase subgroup of the enolase superfamily catalyzing common β-elimination reactions by α-carbon deprotonation of substrates. The structure of VejACI revealed a notable 20s loop region in the capping domain, which can be a highly conserved structural motif in ACI homologs of agar metabolism. By comparing mutant (mVejAC/H300 N) and native VejACI structures, we identified a conformational change of Ile142 in VejACI that causes spatial expansion in the binding pocket. These observations imply that Ile142 and the 20s loop play important roles in enzymatic reactivity and substrate specificity. The structural phylogenetic analysis of the enolase superfamily including ACIs revealed sequential, structural, and functional relationships related to the emergence of novel substrate specificity.
Organizational Affiliation:
Department of Biotechnology, Korea University Graduate School, 5 Anam-ro, Seoungbuk-gu, Seoul, 02841, South Korea; Protein Structure Group, Korea Basic Science Institute, Ochang, Chungbuk, 28119, South Korea.