Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
Sun, Q.Z., Lin, G.F., Li, L.L., Jin, X.T., Huang, L.Y., Zhang, G., Yang, W., Chen, K., Xiang, R., Chen, C., Wei, Y.Q., Lu, G.W., Yang, S.Y.(2017) J Med Chem 60: 6337-6352
- PubMed: 28692292
- DOI: https://doi.org/10.1021/acs.jmedchem.7b00665
- Primary Citation of Related Structures:
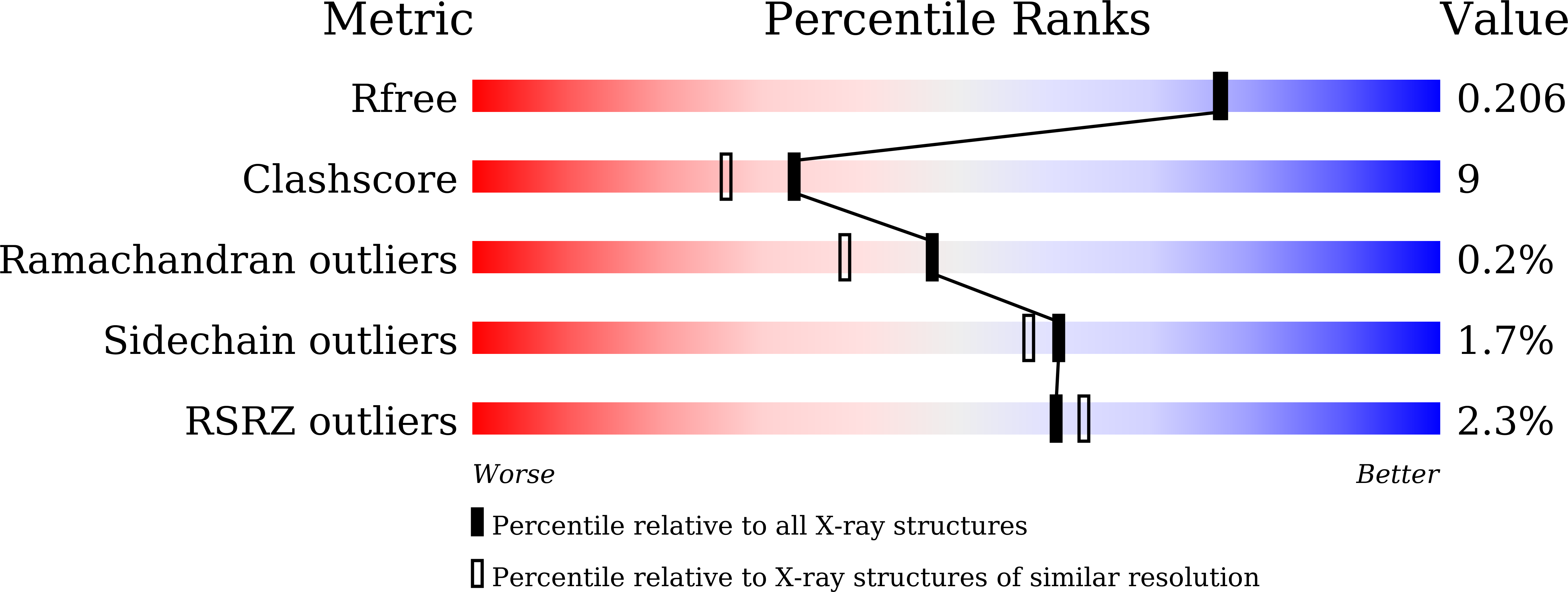
5X8I - PubMed Abstract:
Autophagy inducers represent new promising agents for the treatment of a wide range of medical illnesses. However, safe autophagy inducers for clinical applications are lacking. Inhibition of cdc2-like kinase 1 (CLK1) was recently found to efficiently induce autophagy. Unfortunately, most of the known CLK1 inhibitors have unsatisfactory selectivity. Herein, we report the discovery of a series of new CLK1 inhibitors containing the 1H-[1,2,3]triazolo[4,5-c]quinoline scaffold. Among them, compound 25 was the most potent and selective, with an IC 50 value of 2 nM against CLK1. The crystal structure of CLK1 complexed with compound 25 was solved, and the potency and kinase selectivity of compound 25 were interpreted. Compound 25 was able to induce autophagy in in vitro assays and displayed significant hepatoprotective effects in the acetaminophen (APAP)-induced liver injury mouse model. Collectively, due to its potency and selectivity, compound 25 could be used as a chemical probe or agent in future mechanism-of-action or autophagy-related disease therapy studies.
Organizational Affiliation:
State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University and Collaborative Innovation Center for Biotherapy , Chengdu 610041, P.R. China.