Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG
Luo, Q., Yang, X., Yu, S., Shi, H., Wang, K., Xiao, L., Zhu, G., Sun, C., Li, T., Li, D., Zhang, X., Zhou, M., Huang, Y.(2017) Nat Struct Mol Biol 24: 469-474
- PubMed: 28394325
- DOI: https://doi.org/10.1038/nsmb.3399
- Primary Citation of Related Structures:
5X5Y - PubMed Abstract:
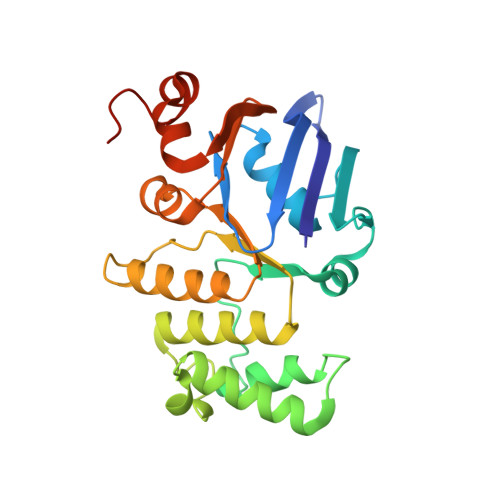
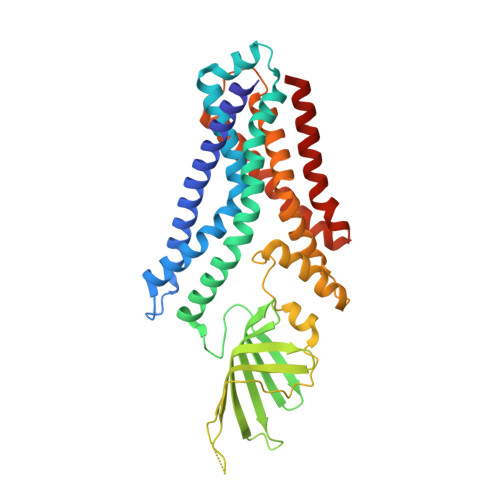
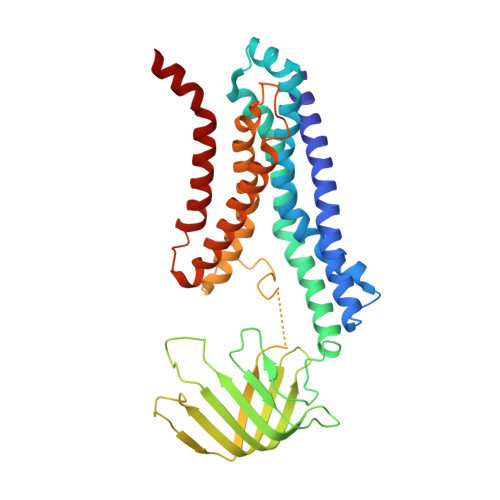
After biosynthesis, bacterial lipopolysaccharides (LPS) are transiently anchored to the outer leaflet of the inner membrane (IM). The ATP-binding cassette (ABC) transporter LptB 2 FG extracts LPS molecules from the IM and transports them to the outer membrane. Here we report the crystal structure of nucleotide-free LptB 2 FG from Pseudomonas aeruginosa. The structure reveals that lipopolysaccharide transport proteins LptF and LptG each contain a transmembrane domain (TMD), a periplasmic β-jellyroll-like domain and a coupling helix that interacts with LptB on the cytoplasmic side. The LptF and LptG TMDs form a large outward-facing V-shaped cavity in the IM. Mutational analyses suggest that LPS may enter the central cavity laterally, via the interface of the TMD domains of LptF and LptG, and is expelled into the β-jellyroll-like domains upon ATP binding and hydrolysis by LptB. These studies suggest a mechanism for LPS extraction by LptB 2 FG that is distinct from those of classical ABC transporters that transport substrates across the IM.
Organizational Affiliation:
National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.