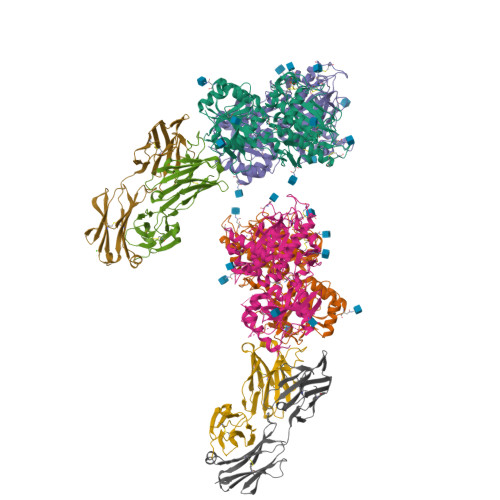
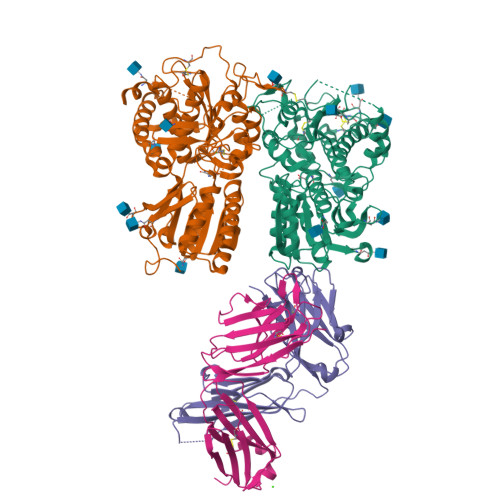
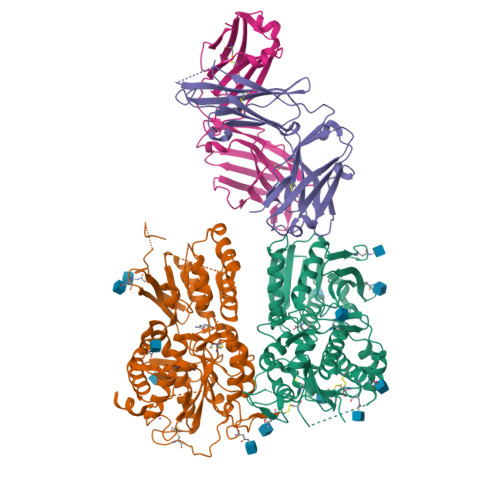
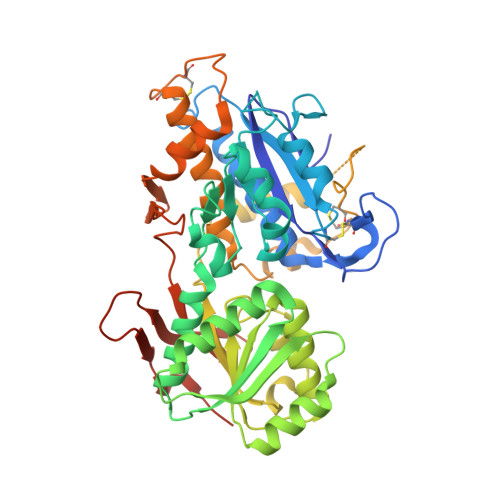
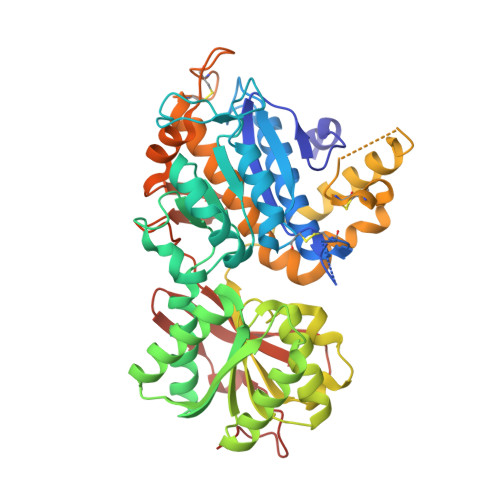
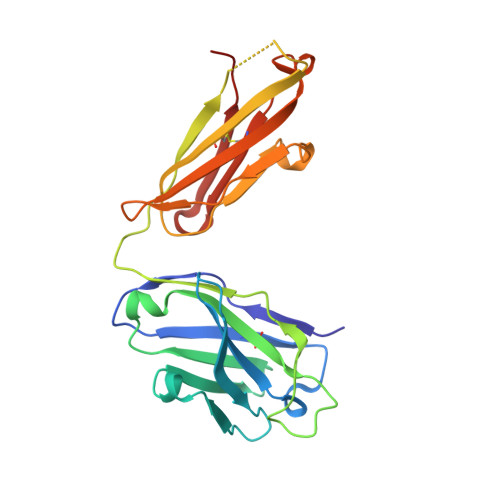
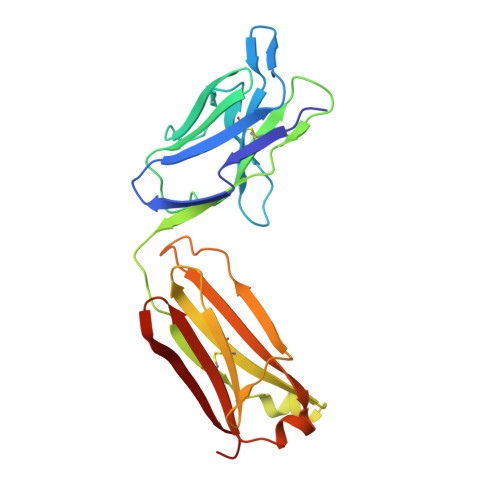
Structural basis for perception of diverse chemical substances by T1r taste receptors
Nuemket, N., Yasui, N., Kusakabe, Y., Nomura, Y., Atsumi, N., Akiyama, S., Nango, E., Kato, Y., Kaneko, M.K., Takagi, J., Hosotani, M., Yamashita, A.(2017) Nat Commun 8: 15530-15530
- PubMed: 28534491
- DOI: https://doi.org/10.1038/ncomms15530
- Primary Citation of Related Structures:
5X2M, 5X2N, 5X2O, 5X2P, 5X2Q - PubMed Abstract:
The taste receptor type 1 (T1r) family perceives 'palatable' tastes. These receptors function as T1r2-T1r3 and T1r1-T1r3 heterodimers to recognize a wide array of sweet and umami (savory) tastes in sugars and amino acids. Nonetheless, it is unclear how diverse tastes are recognized by so few receptors. Here we present crystal structures of the extracellular ligand-binding domains (LBDs), the taste recognition regions of the fish T1r2-T1r3 heterodimer, bound to different amino acids. The ligand-binding pocket in T1r2LBD is rich in aromatic residues, spacious and accommodates hydrated percepts. Biophysical studies show that this binding site is characterized by a broad yet discriminating chemical recognition, contributing for the particular trait of taste perception. In contrast, the analogous pocket in T1r3LBD is occupied by a rather loosely bound amino acid, suggesting that the T1r3 has an auxiliary role. Overall, we provide a structural basis for understanding the chemical perception of taste receptors.
Organizational Affiliation:
Laboratory of Structural Biology, Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, 1-1-1, Tsushima-naka, Okayama 700-8530, Japan.