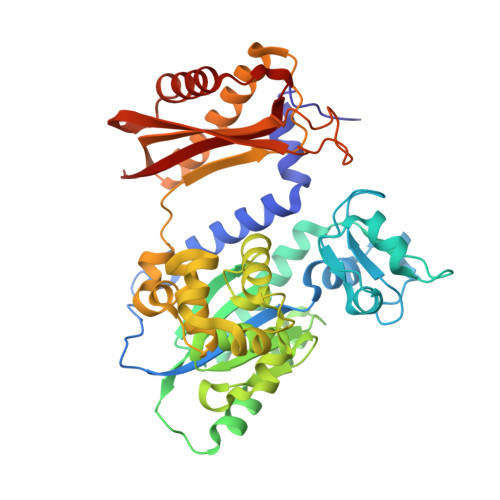
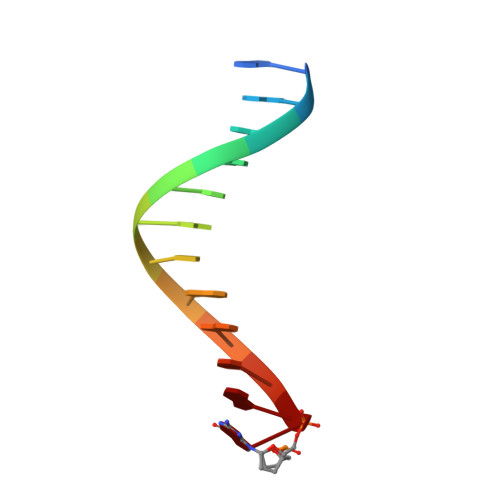
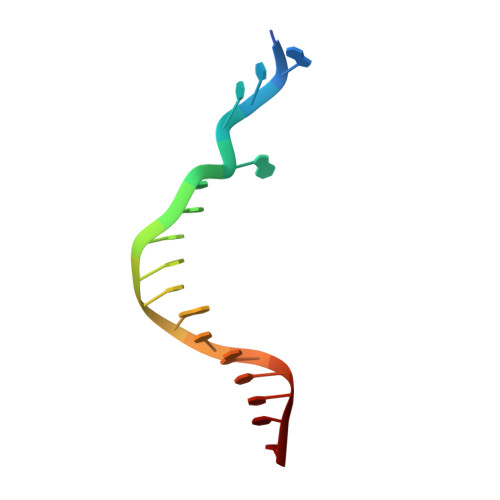
Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Rechkoblit, O., Kolbanovskiy, A., Landes, H., Geacintov, N.E., Aggarwal, A.K.(2017) Nat Commun 8: 965-965
- PubMed: 29042535
- DOI: https://doi.org/10.1038/s41467-017-01013-5
- Primary Citation of Related Structures:
5WM1, 5WM8, 5WMB - PubMed Abstract:
Benzo[a]pyrene (BP) is a carcinogen in cigarette smoke which, after metabolic activation, can react with the exocyclic N 2 amino group of guanine to generate four stereoisomeric BP-N 2 -dG adducts. Rev1 is unique among translesion synthesis DNA polymerases in employing a protein-template-directed mechanism of DNA synthesis opposite undamaged and damaged guanine. Here we report high-resolution structures of yeast Rev1 with three BP-N 2 -dG adducts, namely the 10S (+)-trans-BP-N 2 -dG, 10R (+)-cis-BP-N 2 -dG, and 10S ( - )-cis-BP-N 2 -dG. Surprisingly, in all three structures, the bulky and hydrophobic BP pyrenyl residue is entirely solvent-exposed in the major groove of the DNA. This is very different from the adduct alignments hitherto observed in free or protein-bound DNA. All complexes are well poised for dCTP insertion. Our structures provide a view of cis-BP-N 2 -dG adducts in a DNA polymerase active site, and offer a basis for understanding error-free replication of the BP-derived stereoisomeric guanine adducts.Benzo[a]pyrene (BP) is a carcinogen in cigarette smoke that upon metabolic activation reacts with guanine. Here, the authors present the structures of the translesion DNA synthesis polymerase Rev1 in complex with three of the four possible stereoisomeric BP-N 2 -dG adducts, which gives insights how Rev1 achieves error-free replication.
Organizational Affiliation:
Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, Box 1677, 1425 Madison Avenue, New York, NY, 10029, USA. olga.rechkoblit@mssm.edu.