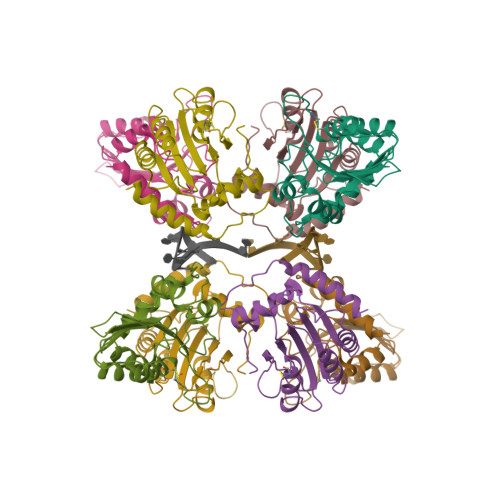
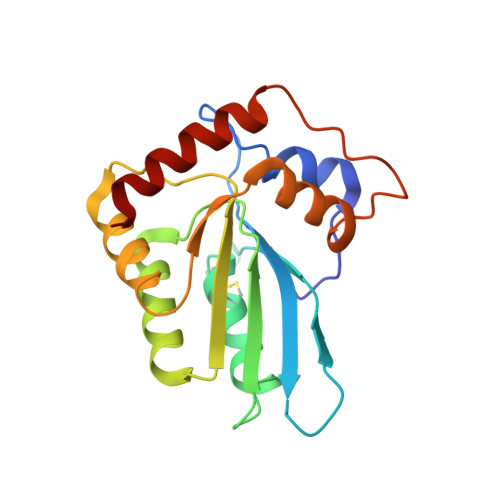
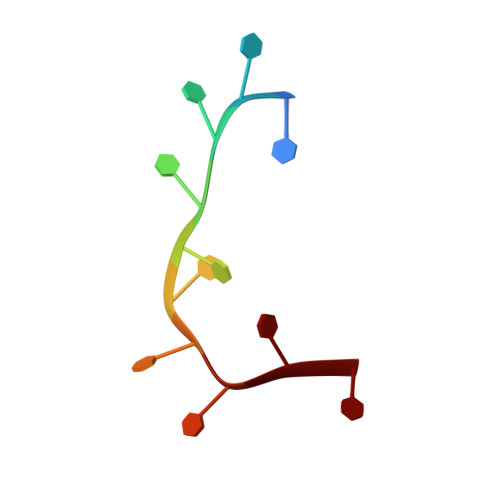
Molecular Interactions of a DNA Modifying Enzyme APOBEC3F Catalytic Domain with a Single-Stranded DNA.
Fang, Y., Xiao, X., Li, S.X., Wolfe, A., Chen, X.S.(2018) J Mol Biology 430: 87-101
- PubMed: 29191651
- DOI: https://doi.org/10.1016/j.jmb.2017.11.007
- Primary Citation of Related Structures:
5W2M - PubMed Abstract:
The single-stranded DNA (ssDNA) cytidine deaminase APOBEC3F (A3F) deaminates cytosine (C) to uracil (U) and is a known restriction factor of HIV-1. Its C-terminal catalytic domain (CD2) alone is capable of binding single-stranded nucleic acids and is important for deamination. However, little is known about how the CD2 interacts with ssDNA. Here we report a crystal structure of A3F-CD2 in complex with a 10-nucleotide ssDNA composed of poly-thymine, which reveals a novel positively charged nucleic acid binding site distal to the active center that plays a key role in substrate DNA binding and catalytic activity. Lysine and tyrosine residues within this binding site interact with the ssDNA, and mutating these residues dramatically impairs both ssDNA binding and catalytic activity. This binding site is not conserved in APOBEC3G (A3G), which may explain differences in ssDNA-binding characteristics between A3F-CD2 and A3G-CD2. In addition, we observed an alternative Zn-coordination conformation around the active center. These findings reveal the structural relationships between nucleic acid interactions and catalytic activity of A3F.
Organizational Affiliation:
Molecular and Computational Biology, Departments of Biological Sciences and Chemistry, University of Southern California, Los Angeles, CA 90089, USA; 161 Hospital of PLA, Wuhan, 430012, China; Department of Clinical Microbiology and Immunology of Southwest Hospital, Third Military Medical University, Chongqing 400038, China.