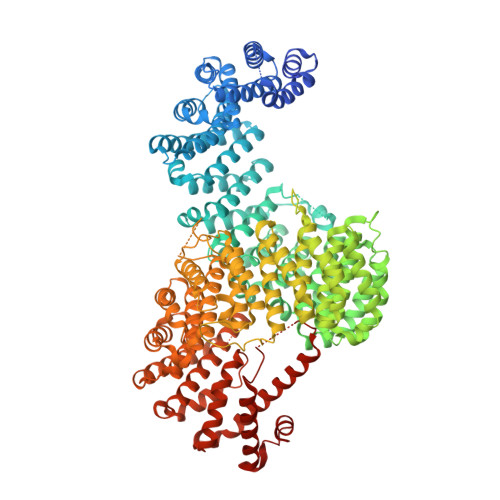
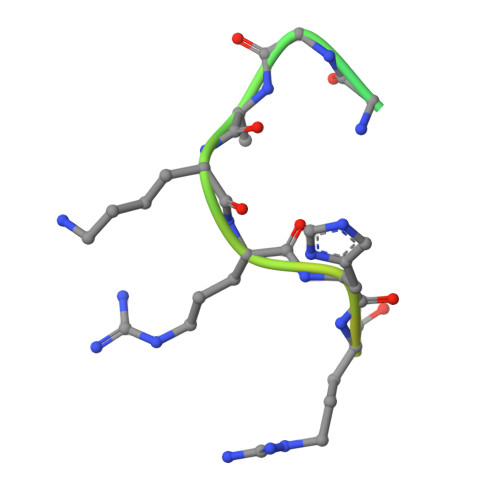
Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123.
An, S., Yoon, J., Kim, H., Song, J.J., Cho, U.S.(2017) Elife 6
- PubMed: 29035199
- DOI: https://doi.org/10.7554/eLife.30244
- Primary Citation of Related Structures:
5VCH, 5VE8, 5W0V - PubMed Abstract:
Kap123, a major karyopherin protein of budding yeast, recognizes the nuclear localization signals (NLSs) of cytoplasmic histones H3 and H4 and translocates them into the nucleus during DNA replication. Mechanistic questions include H3- and H4-NLS redundancy toward Kap123 and the role of the conserved diacetylation of cytoplasmic H4 (K5ac and K12ac) in Kap123-mediated histone nuclear translocation. Here, we report crystal structures of full-length Kluyveromyces lactis Kap123 alone and in complex with H3- and H4-NLSs. Structures reveal the unique feature of Kap123 that possesses two discrete lysine-binding pockets for NLS recognition. Structural comparison illustrates that H3- and H4-NLSs share at least one of two lysine-binding pockets, suggesting that H3- and H4-NLSs are mutually exclusive. Additionally, acetylation of key lysine residues at NLS, particularly H4-NLS diacetylation, weakens the interaction with Kap123. These data support that cytoplasmic histone H4 diacetylation weakens the Kap123-H4-NLS interaction thereby facilitating histone Kap123-H3-dependent H3:H4/Asf1 complex nuclear translocation.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan Medical School, Michigan, United States.