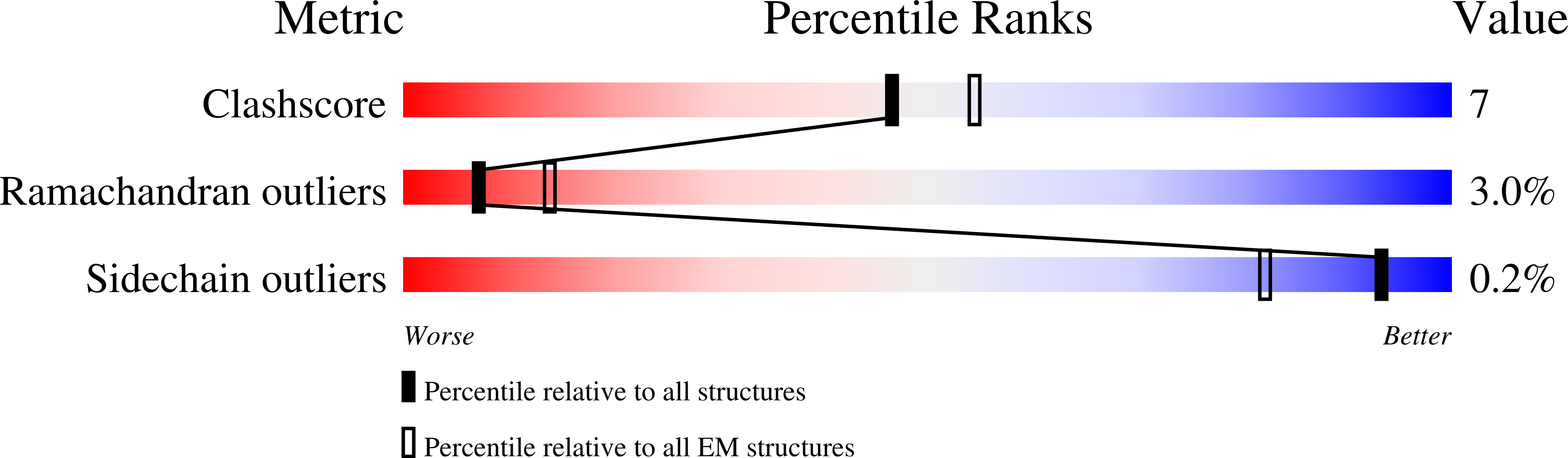Lis1 Has Two Opposing Modes of Regulating Cytoplasmic Dynein.
DeSantis, M.E., Cianfrocco, M.A., Htet, Z.M., Tran, P.T., Reck-Peterson, S.L., Leschziner, A.E.(2017) Cell 170: 1197-1208.e12
- PubMed: 28886386
- DOI: https://doi.org/10.1016/j.cell.2017.08.037
- Primary Citation of Related Structures:
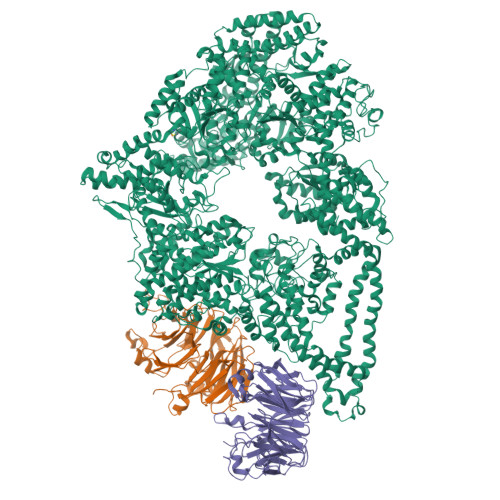
5VH9, 5VLJ - PubMed Abstract:
Regulation is central to the functional versatility of cytoplasmic dynein, a motor involved in intracellular transport, cell division, and neurodevelopment. Previous work established that Lis1, a conserved regulator of dynein, binds to its motor domain and induces a tight microtubule-binding state in dynein. The work we present here-a combination of biochemistry, single-molecule assays, and cryoelectron microscopy-led to the surprising discovery that Lis1 has two opposing modes of regulating dynein, being capable of inducing both low and high affinity for the microtubule. We show that these opposing modes depend on the stoichiometry of Lis1 binding to dynein and that this stoichiometry is regulated by the nucleotide state of dynein's AAA3 domain. The low-affinity state requires Lis1 to also bind to dynein at a novel conserved site, mutation of which disrupts Lis1's function in vivo. We propose a new model for the regulation of dynein by Lis1.
Organizational Affiliation:
Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92093, USA.