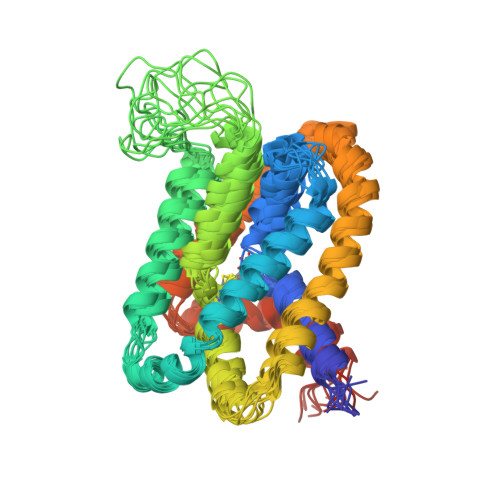
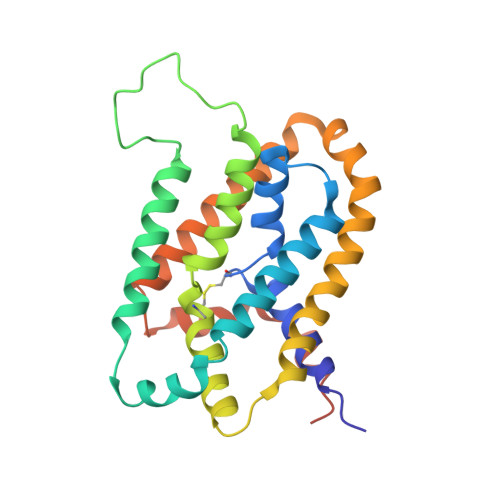
Solution structure and elevator mechanism of the membrane electron transporter CcdA.
Zhou, Y., Bushweller, J.H.(2018) Nat Struct Mol Biol 25: 163-169
- PubMed: 29379172
- DOI: https://doi.org/10.1038/s41594-018-0022-z
- Primary Citation of Related Structures:
5VKV - PubMed Abstract:
Membrane oxidoreductase CcdA plays a central role in supplying reducing equivalents from the bacterial cytoplasm to the envelope. It transports electrons across the membrane using a single pair of cysteines by a mechanism that has not yet been elucidated. Here we report an NMR structure of the Thermus thermophilus CcdA (TtCcdA) in an oxidized and outward-facing state. CcdA consists of two inverted structural repeats of three transmembrane helices (2 × 3-TM). We computationally modeled and experimentally validated an inward-facing state, which suggests that CcdA uses an elevator-type movement to shuttle the reactive cysteines across the membrane. CcdA belongs to the LysE superfamily, and thus its structure may be relevant to other LysE clan transporters. Structure comparisons of CcdA, semiSWEET, Pnu, and major facilitator superfamily (MFS) transporters provide insights into membrane transporter architecture and mechanism.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA, USA. yz8w@virginia.edu.