La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs.
Lahr, R.M., Fonseca, B.D., Ciotti, G.E., Al-Ashtal, H.A., Jia, J.J., Niklaus, M.R., Blagden, S.P., Alain, T., Berman, A.J.(2017) Elife 6
- PubMed: 28379136
- DOI: https://doi.org/10.7554/eLife.24146
- Primary Citation of Related Structures:
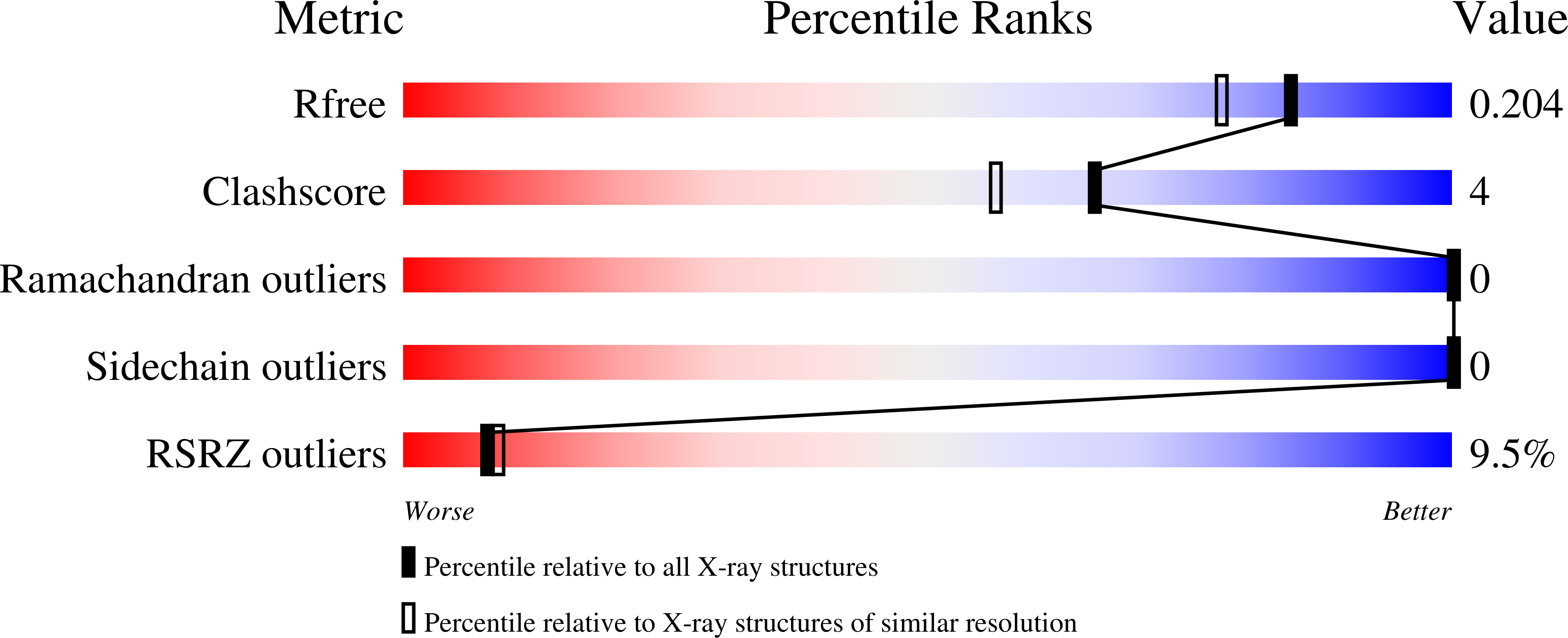
5V4R, 5V7C, 5V87 - PubMed Abstract:
The 5'terminal oligopyrimidine (5'TOP) motif is a cis -regulatory RNA element located immediately downstream of the 7-methylguanosine [m 7 G] cap of TOP mRNAs, which encode ribosomal proteins and translation factors. In eukaryotes, this motif coordinates the synchronous and stoichiometric expression of the protein components of the translation machinery. La-related protein 1 (LARP1) binds TOP mRNAs, regulating their stability and translation. We present crystal structures of the human LARP1 DM15 region in complex with a 5'TOP motif, a cap analog (m 7 GTP), and a capped cytidine (m 7 GpppC), resolved to 2.6, 1.8 and 1.7 Å, respectively. Our binding, competition, and immunoprecipitation data corroborate and elaborate on the mechanism of 5'TOP motif binding by LARP1. We show that LARP1 directly binds the cap and adjacent 5'TOP motif of TOP mRNAs, effectively impeding access of eIF4E to the cap and preventing eIF4F assembly. Thus, LARP1 is a specialized TOP mRNA cap-binding protein that controls ribosome biogenesis.
Organizational Affiliation:
Department of Biological Sciences, University of Pittsburgh, Pittsburgh, United States.