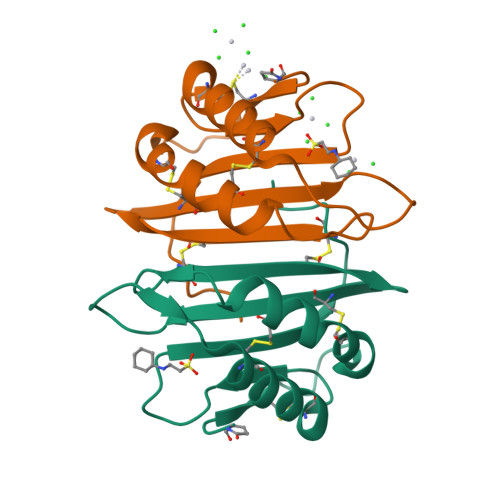
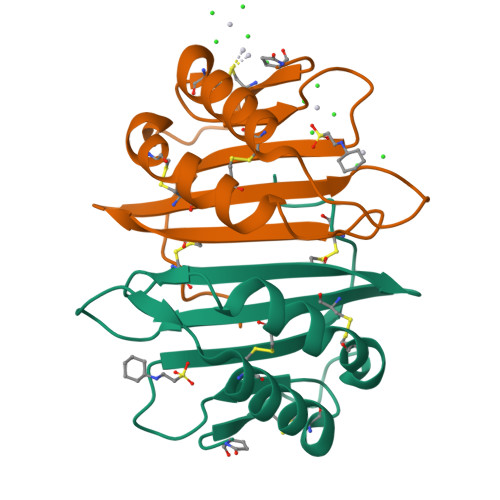
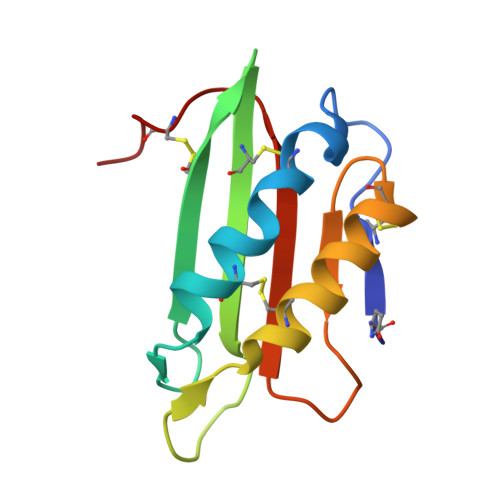
Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Zhang, P., Li, K., Yang, G., Xia, C., Polston, J.E., Li, G., Li, S., Lin, Z., Yang, L.J., Bruner, S.D., Ding, Y.(2017) Proc Natl Acad Sci U S A 114: 8980-8985
- PubMed: 28784797
- DOI: https://doi.org/10.1073/pnas.1706894114
- Primary Citation of Related Structures:
5V6I, 5V6J - PubMed Abstract:
Glycans possess significant chemical diversity; glycan binding proteins (GBPs) recognize specific glycans to translate their structures to functions in various physiological and pathological processes. Therefore, the discovery and characterization of novel GBPs and characterization of glycan-GBP interactions are significant to provide potential targets for therapeutic intervention of many diseases. Here, we report the biochemical, functional, and structural characterization of a 130-amino-acid protein, Y3, from the mushroom Coprinus comatus Biochemical studies of recombinant Y3 from a yeast expression system demonstrated the protein is a unique GBP. Additionally, we show that Y3 exhibits selective and potent cytotoxicity toward human T-cell leukemia Jurkat cells compared with a panel of cancer cell lines via inducing caspase-dependent apoptosis. Screening of a glycan array demonstrated GalNAcβ1-4(Fucα1-3)GlcNAc (LDNF) as a specific Y3-binding ligand. To provide a structural basis for function, the crystal structure was solved to a resolution of 1.2 Å, revealing a single-domain αβα-sandwich motif. Two monomers were dimerized to form a large 10-stranded, antiparallel β-sheet flanked by α-helices on each side, representing a unique oligomerization mode among GBPs. A large glycan binding pocket extends into the dimeric interface, and docking of LDNF identified key residues for glycan interactions. Disruption of residues predicted to be involved in LDNF/Y3 interactions resulted in the significant loss of binding to Jurkat T-cells and severely impaired their cytotoxicity. Collectively, these results demonstrate Y3 to be a GBP with selective cytotoxicity toward human T-cell leukemia cells and indicate its potential use in cancer diagnosis and treatment.
Organizational Affiliation:
Department of Medicinal Chemistry, Center for Natural Products, Drug Discovery and Development, University of Florida, Gainesville, FL 32610.