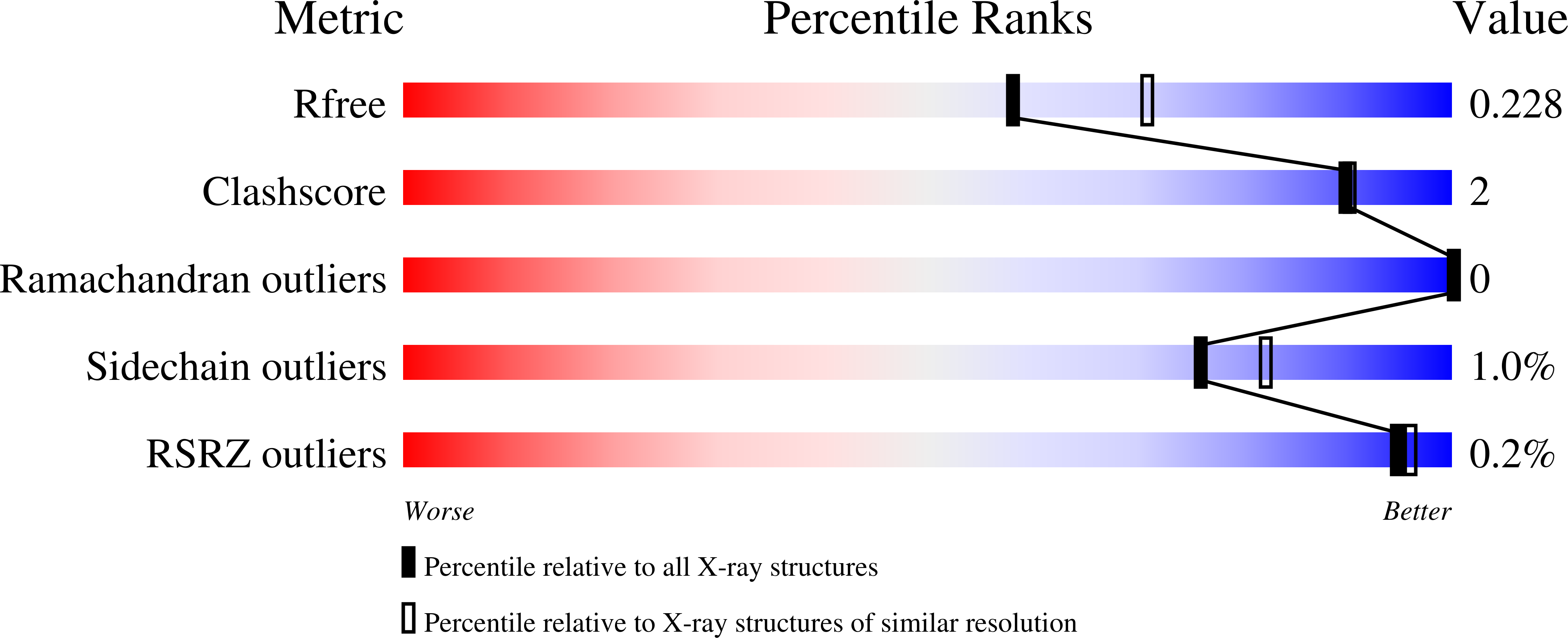
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
Korasick, D.A., Singh, H., Pemberton, T.A., Luo, M., Dhatwalia, R., Tanner, J.J.(2017) FEBS J 284: 3029-3049
- PubMed: 28710792
- DOI: https://doi.org/10.1111/febs.14165
- Primary Citation of Related Structures:
5UR2 - PubMed Abstract:
Many enzymes form homooligomers, yet the functional significance of self-association is seldom obvious. Herein, we examine the connection between oligomerization and catalytic function for proline utilization A (PutA) enzymes. PutAs are bifunctional enzymes that catalyze both reactions of proline catabolism. Type A PutAs are the smallest members of the family, possessing a minimal domain architecture consisting of N-terminal proline dehydrogenase and C-terminal l-glutamate-γ-semialdehyde dehydrogenase modules. Type A PutAs form domain-swapped dimers, and in one case (Bradyrhizobium japonicum PutA), two of the dimers assemble into a ring-shaped tetramer. Whereas the dimer has a clear role in substrate channeling, the functional significance of the tetramer is unknown. To address this question, we performed structural studies of four-type A PutAs from two clades of the PutA tree. The crystal structure of Bdellovibrio bacteriovorus PutA covalently inactivated by N-propargylglycine revealed a fold and substrate-channeling tunnel similar to other PutAs. Small-angle X-ray scattering (SAXS) and analytical ultracentrifugation indicated that Bdellovibrio PutA is dimeric in solution, in contrast to the prediction from crystal packing of a stable tetrameric assembly. SAXS studies of two other type A PutAs from separate clades also suggested that the dimer predominates in solution. To assess whether the tetramer of B. japonicum PutA is necessary for catalytic function, a hot spot disruption mutant that cleanly produces dimeric protein was generated. The dimeric variant exhibited kinetic parameters similar to the wild-type enzyme. These results implicate the domain-swapped dimer as the core structural and functional unit of type A PutAs. Proline dehydrogenase (EC 1.5.5.2); l-glutamate-γ-semialdehyde dehydrogenase (EC 1.2.1.88). The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession number 5UR2. The SAXS data have been deposited in the SASBDB under the following accession codes: SASDCP3 (BbPutA), SASDCQ3 (DvPutA 1.5 mg·mL -1 ), SASDCX3 (DvPutA 3.0 mg·mL -1 ), SASDCY3 (DvPutA 4.5 mg·mL -1 ), SASDCR3 (LpPutA 3.0 mg·mL -1 ), SASDCV3 (LpPutA 5.0 mg·mL -1 ), SASDCW3 (LpPutA 8.0 mg·mL -1 ), SASDCS3 (BjPutA 2.3 mg·mL -1 ), SASDCT3 (BjPutA 4.7 mg·mL -1 ), SASDCU3 (BjPutA 7.0 mg·mL -1 ), SASDCZ3 (R51E 2.3 mg·mL -1 ), SASDC24 (R51E 4.7 mg·mL -1 ), SASDC34 (R51E 7.0 mg·mL -1 ).
Organizational Affiliation:
Department of Biochemistry, University of Missouri, Columbia, MO, USA.