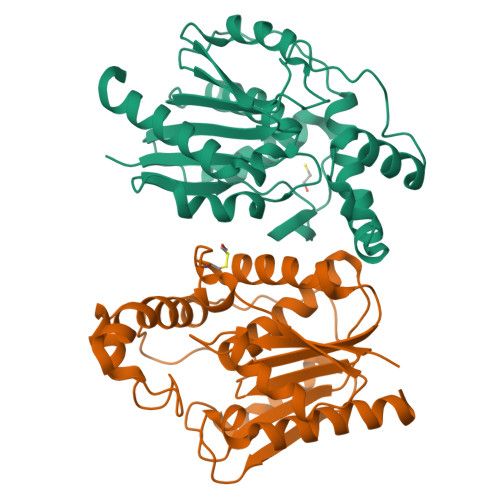
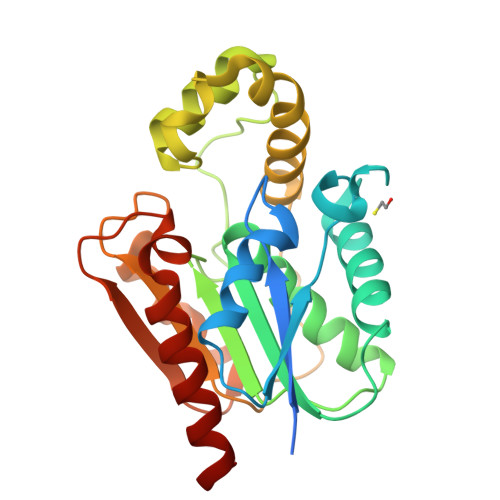
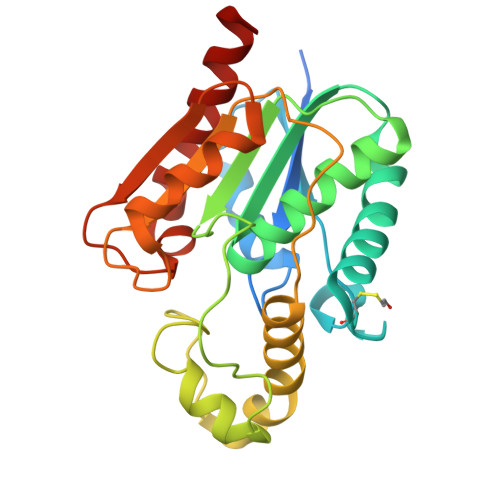
Structure and Functional Analysis of ClbQ, an Unusual Intermediate-Releasing Thioesterase from the Colibactin Biosynthetic Pathway.
Guntaka, N.S., Healy, A.R., Crawford, J.M., Herzon, S.B., Bruner, S.D.(2017) ACS Chem Biol 12: 2598-2608
- PubMed: 28846367
- DOI: https://doi.org/10.1021/acschembio.7b00479
- Primary Citation of Related Structures:
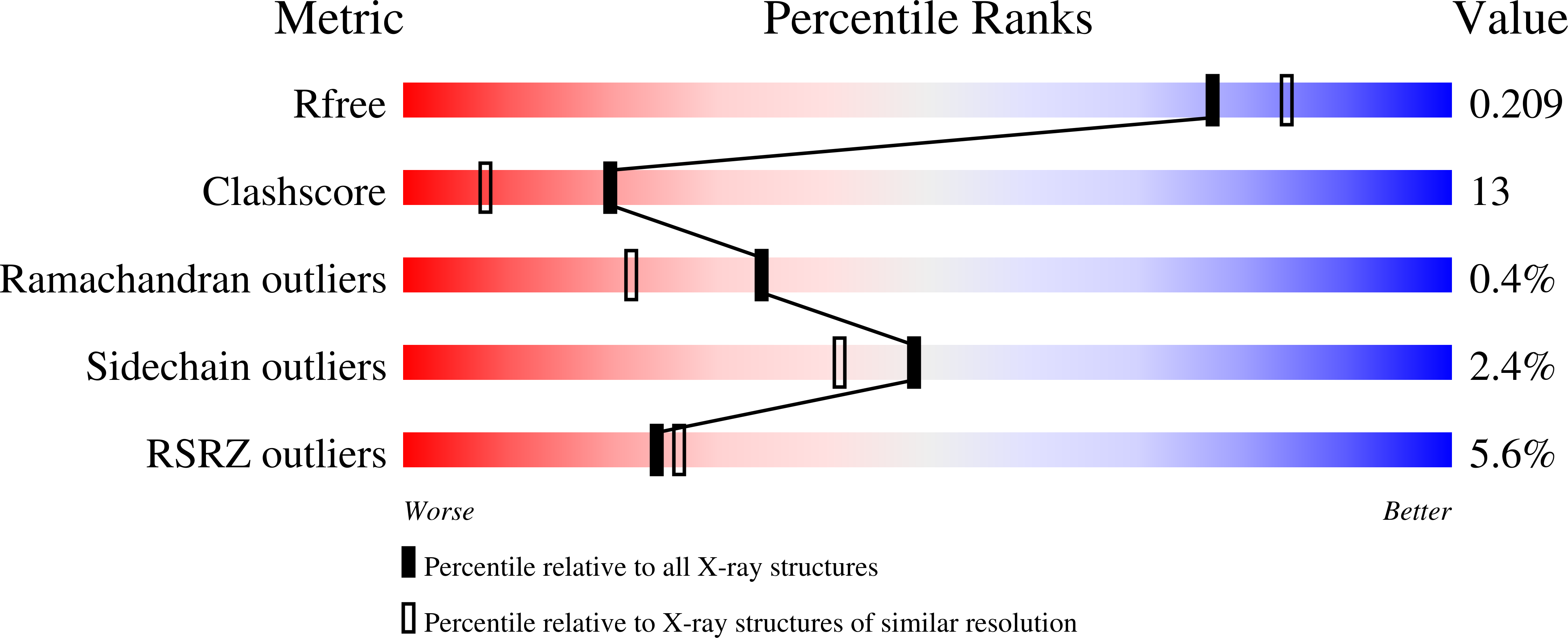
5UGZ - PubMed Abstract:
Colibactin is a genotoxic hybrid nonribosomal peptide/polyketide secondary metabolite produced by various pathogenic and probiotic bacteria residing in the human gut. The presence of colibactin metabolites has been correlated to colorectal cancer formation in several studies. The specific function of many gene products in the colibactin gene cluster can be predicted. However, the role of ClbQ, a type II editing thioesterase, has not been established. The importance of ClbQ has been demonstrated by genetic deletions that abolish colibactin cytotoxic activity, and recent studies suggest an atypical role in releasing pathway intermediates from the assembly line. Here we report the 2.0 Å crystal structure and biochemical characterization of ClbQ. Our data reveal that ClbQ exhibits greater catalytic efficiency toward acyl-thioester substrates as compared to precolibactin intermediates and does not discriminate among carrier proteins. Cyclized pyridone-containing colibactins, which are off-pathway derivatives, are not viable substrates for ClbQ, while linear precursors are, supporting a role of ClbQ in facilitating the promiscuous off-loading of premature precolibactin metabolites and novel insights into colibactin biosynthesis.
Organizational Affiliation:
Department of Chemistry, University of Florida , Gainesville, Florida 32611, United States.