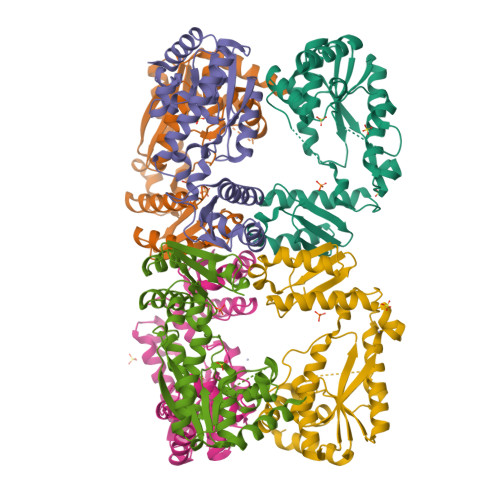
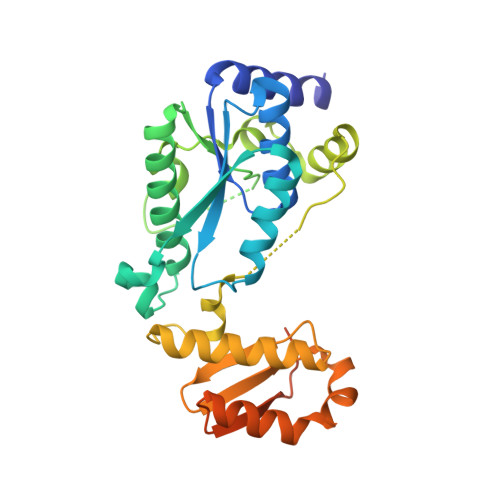
Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Fellner, M., Desguin, B., Hausinger, R.P., Hu, J.(2017) Proc Natl Acad Sci U S A 114: 9074-9079
- PubMed: 28784764
- DOI: https://doi.org/10.1073/pnas.1704967114
- Primary Citation of Related Structures:
5UDQ, 5UDR, 5UDS, 5UDT, 5UDU, 5UDV, 5UDW, 5UDX, 5UNM - PubMed Abstract:
The lar operon in Lactobacillus plantarum encodes five Lar proteins (LarA/B/C/D/E) that collaboratively synthesize and incorporate a niacin-derived Ni-containing cofactor into LarA, an Ni-dependent lactate racemase. Previous studies have established that two molecules of LarE catalyze successive thiolation reactions by donating the sulfur atom of their exclusive cysteine residues to the substrate. However, the catalytic mechanism of this very unusual sulfur-sacrificing reaction remains elusive. In this work, we present the crystal structures of LarE in ligand-free and several ligand-bound forms, demonstrating that LarE is a member of the N-type ATP pyrophosphatase (PPase) family with a conserved N-terminal ATP PPase domain and a unique C-terminal domain harboring the putative catalytic site. Structural analysis, combined with structure-guided mutagenesis, leads us to propose a catalytic mechanism that establishes LarE as a paradigm for sulfur transfer through sacrificing its catalytic cysteine residue.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824.