A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants.
Zhu, Q., McLellan, J.S., Kallewaard, N.L., Ulbrandt, N.D., Palaszynski, S., Zhang, J., Moldt, B., Khan, A., Svabek, C., McAuliffe, J.M., Wrapp, D., Patel, N.K., Cook, K.E., Richter, B.W.M., Ryan, P.C., Yuan, A.Q., Suzich, J.A.(2017) Sci Transl Med 9
- PubMed: 28469033
- DOI: https://doi.org/10.1126/scitranslmed.aaj1928
- Primary Citation of Related Structures:
5UDC, 5UDD, 5UDE - PubMed Abstract:
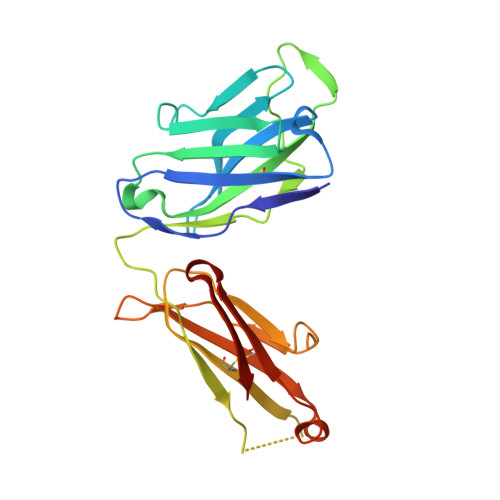
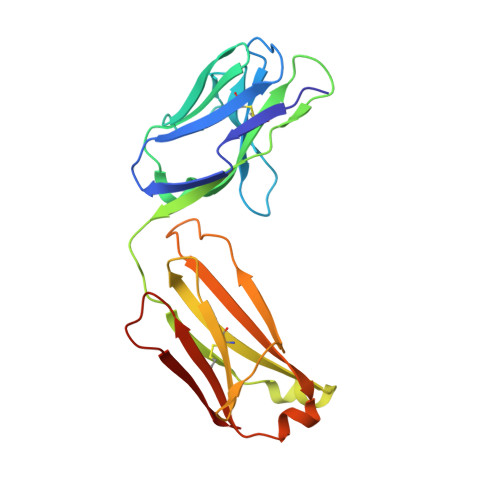
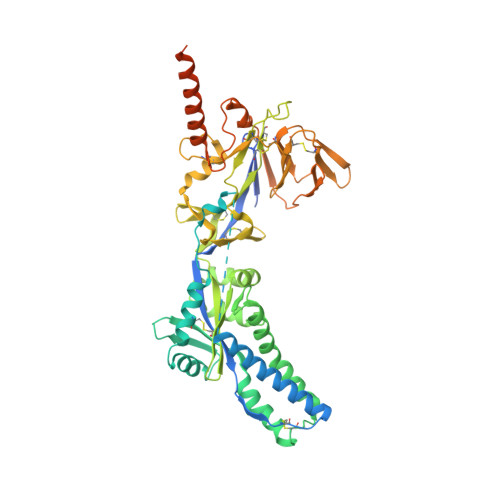
Prevention of respiratory syncytial virus (RSV) illness in all infants is a major public health priority. However, no vaccine is currently available to protect this vulnerable population. Palivizumab, the only approved agent for RSV prophylaxis, is limited to high-risk infants, and the cost associated with the requirement for dosing throughout the RSV season makes its use impractical for all infants. We describe the development of a monoclonal antibody as potential RSV prophylaxis for all infants with a single intramuscular dose. MEDI8897*, a highly potent human antibody, was optimized from antibody D25, which targets the prefusion conformation of the RSV fusion (F) protein. Crystallographic analysis of Fab in complex with RSV F from subtypes A and B reveals that MEDI8897* binds a highly conserved epitope. MEDI8897* neutralizes a diverse panel of RSV A and B strains with >50-fold higher activity than palivizumab. At similar serum concentrations, prophylactic administration of MEDI8897* was ninefold more potent than palivizumab at reducing pulmonary viral loads by >3 logs in cotton rats infected with either RSV A or B subtypes. MEDI8897 was generated by the introduction of triple amino acid substitutions (YTE) into the Fc domain of MEDI8897*, which led to more than threefold increased half-life in cynomolgus monkeys compared to non-YTE antibody. Considering the pharmacokinetics of palivizumab in infants, which necessitates five monthly doses for protection during an RSV season, the high potency and extended half-life of MEDI8897 support its development as a cost-effective option to protect all infants from RSV disease with once-per-RSV-season dosing in the clinic.
Organizational Affiliation:
Department of Infectious Disease, MedImmune LLC, One MedImmune Way, Gaithersburg, MD 20878, USA. suzichj@medimmune.com zhuq@medimmune.com.