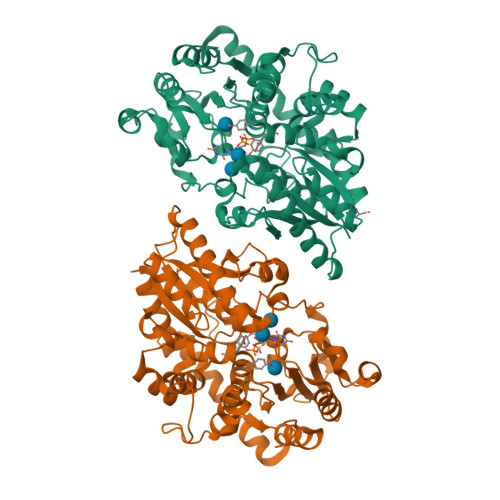
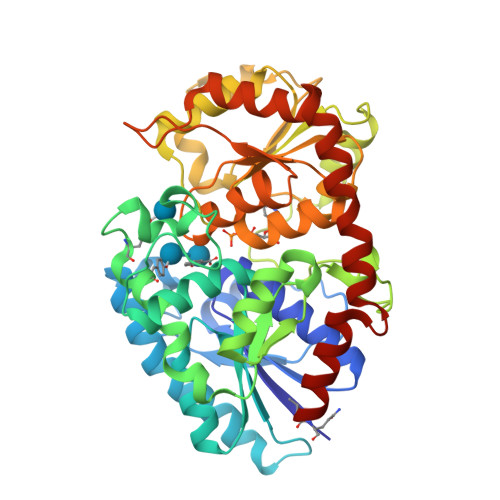
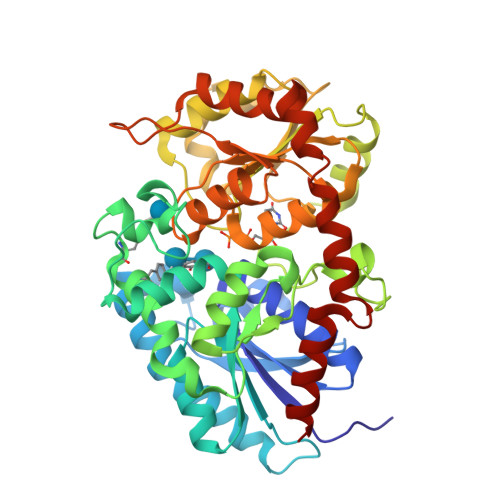
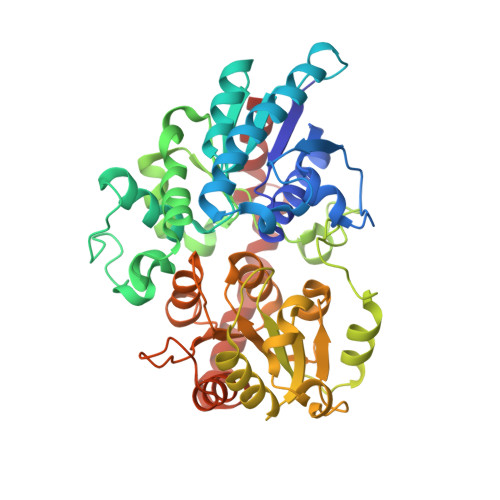
Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana.
George Thompson, A.M., Iancu, C.V., Neet, K.E., Dean, J.V., Choe, J.Y.(2017) Sci Rep 7: 46629-46629
- PubMed: 28425481
- DOI: https://doi.org/10.1038/srep46629
- Primary Citation of Related Structures:
5U6M, 5U6N, 5U6S, 5V2J, 5V2K - PubMed Abstract:
Salicylic acid (SA) is a signaling molecule utilized by plants in response to various stresses. Through conjugation with small organic molecules such as glucose, an inactive form of SA is generated which can be transported into and stored in plant vacuoles. In the model organism Arabidopsis thaliana, SA glucose conjugates are formed by two homologous enzymes (UGT74F1 and UGT74F2) that transfer glucose from UDP-glucose to SA. Despite being 77% identical and with conserved active site residues, these enzymes catalyze the formation of different products: UGT74F1 forms salicylic acid glucoside (SAG), while UGT74F2 forms primarily salicylic acid glucose ester (SGE). The position of the glucose on the aglycone determines how SA is stored, further metabolized, and contributes to a defense response. We determined the crystal structures of the UGT74F2 wild-type and T15S mutant enzymes, in different substrate/product complexes. On the basis of the crystal structures and the effect on enzyme activity of mutations in the SA binding site, we propose the catalytic mechanism of SGE and SAG formation and that SA binds to the active site in two conformations, with each enzyme selecting a certain binding mode of SA. Additionally, we show that two threonines are key determinants of product specificity.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Rosalind Franklin University of Medicine and Science, The Chicago Medical School, 3333 Green Bay Road, North Chicago, IL 60064, USA.