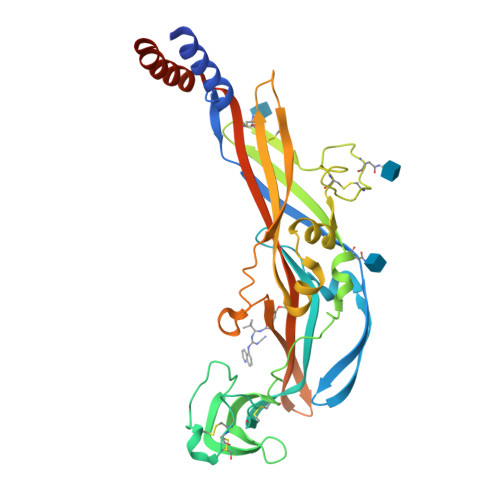
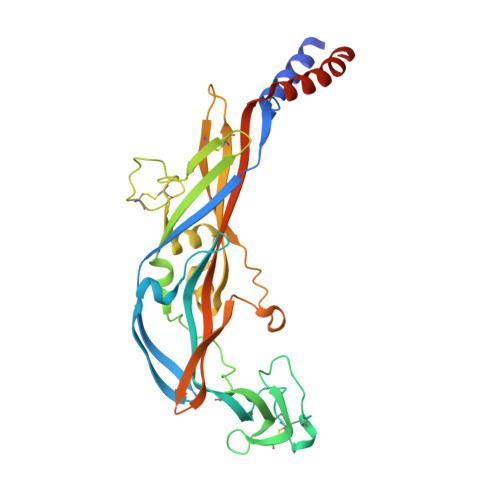
Structural basis for subtype-specific inhibition of the P2X7 receptor.
Karasawa, A., Kawate, T.(2016) Elife 5
- PubMed: 27935479
- DOI: https://doi.org/10.7554/eLife.22153
- Primary Citation of Related Structures:
5U1L, 5U1U, 5U1V, 5U1W, 5U1X, 5U1Y, 5U2H - PubMed Abstract:
The P2X7 receptor is a non-selective cation channel activated by extracellular adenosine triphosphate (ATP). Chronic activation of P2X7 underlies many health problems such as pathologic pain, yet we lack effective antagonists due to poorly understood mechanisms of inhibition. Here we present crystal structures of a mammalian P2X7 receptor complexed with five structurally-unrelated antagonists. Unexpectedly, these drugs all bind to an allosteric site distinct from the ATP-binding pocket in a groove formed between two neighboring subunits. This novel drug-binding pocket accommodates a diversity of small molecules mainly through hydrophobic interactions. Functional assays propose that these compounds allosterically prevent narrowing of the drug-binding pocket and the turret-like architecture during channel opening, which is consistent with a site of action distal to the ATP-binding pocket. These novel mechanistic insights will facilitate the development of P2X7-specific drugs for treating human diseases.
Organizational Affiliation:
Department of Molecular Medicine, Cornell University, Ithaca, United States.