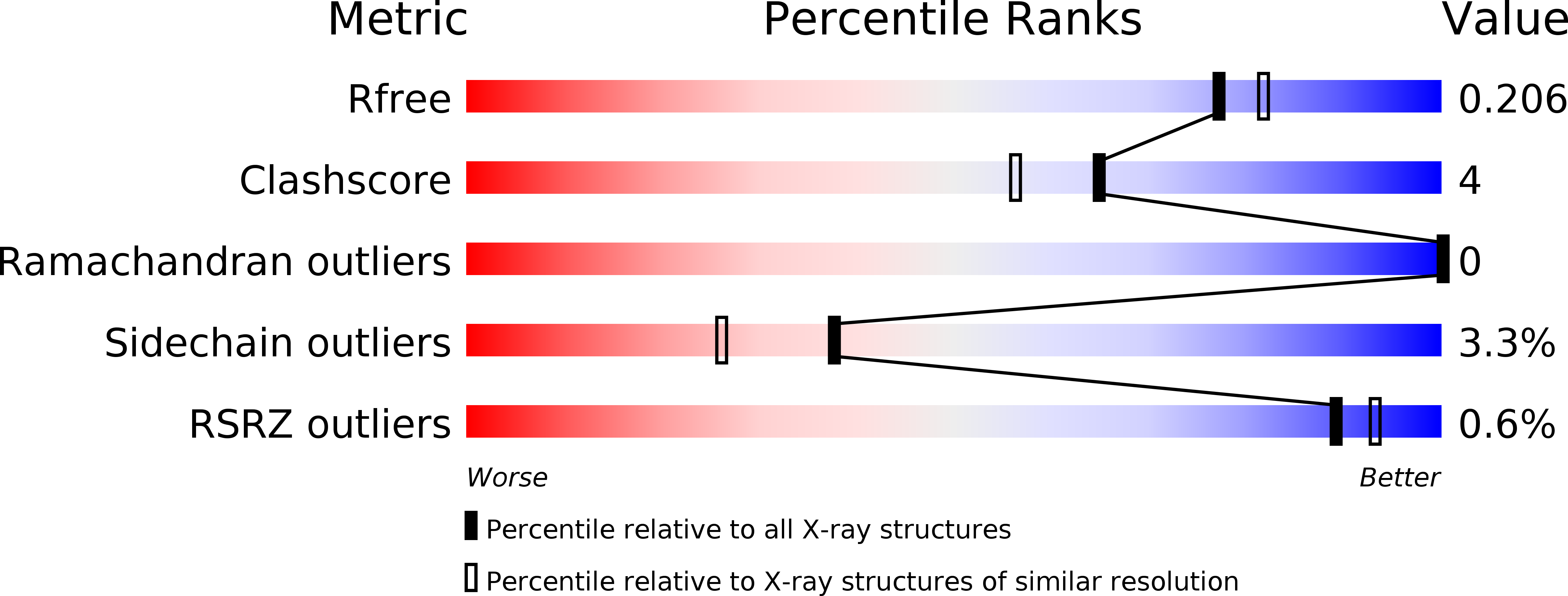
The structure of the Pfp1 protease from the hyperthermophilic archaeon Thermococcus thioreducens in two crystal forms.
Larson, S.B., McPherson, A.(2017) Acta Crystallogr D Struct Biol 73: 749-756
- PubMed: 28876238
- DOI: https://doi.org/10.1107/S2059798317010622
- Primary Citation of Related Structures:
5TW0, 5TXW - PubMed Abstract:
The Pfp1 protease, a cysteine protease of unknown specificity from the hyperthermophilic archaeon Thermococcus thioreducens, was crystallized in two distinctive crystal forms: from concentrated citrate in one case and PEG in the other. X-ray data were collected from both crystal forms at room temperature to about 1.9 Å resolution using a laboratory source and detector, and the structures were solved by molecular replacement using the Pfp1 protease from Pyrococcus horikoshii as the search model. In the T. thioreducens protease structures, Cys18 residues on adjacent molecules in the asymmetric units form intermolecular disulfide bonds, thereby yielding hexamers composed of three cross-linked, quasi-dyad-related dimers with crystallographically exact threefold axes and exhibiting almost exact 32 symmetry. The corresponding residue in P. horikoshii Pfp1 is Tyr18. An individual active site containing Cys100 and His101 also includes a Glu74 residue contributed by a quasi-twofold-related, non-cross-linked subunit. Two catalytic triads are therefore closely juxtaposed about the quasi-twofold axis at the interface of these subunits, and are relatively sequestered within the hexamer cavity. The cysteine in the active site is observed to be oxidized in both of the crystal forms that were studied.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, 530A Steinhaus Hall, Irvine, CA 92697-3900, USA.