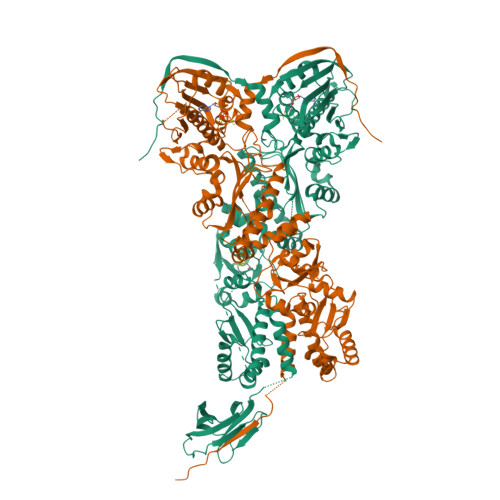
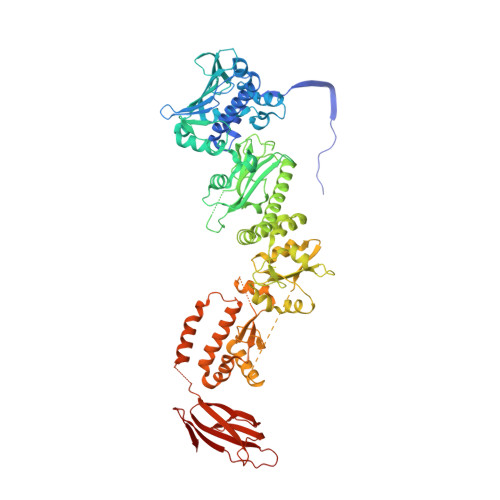
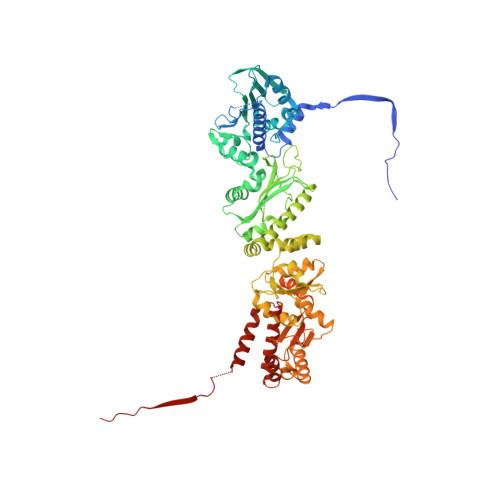
Symmetry broken and rebroken during the ATP hydrolysis cycle of the mitochondrial Hsp90 TRAP1.
Elnatan, D., Betegon, M., Liu, Y., Ramelot, T., Kennedy, M.A., Agard, D.A.(2017) Elife 6
- PubMed: 28742020
- DOI: https://doi.org/10.7554/eLife.25235
- Primary Citation of Related Structures:
5TTH, 5TVU, 5TVW, 5TVX - PubMed Abstract:
Hsp90 is a homodimeric ATP-dependent molecular chaperone that remodels its substrate 'client' proteins, facilitating their folding and activating them for biological function. Despite decades of research, the mechanism connecting ATP hydrolysis and chaperone function remains elusive. Particularly puzzling has been the apparent lack of cooperativity in hydrolysis of the ATP in each protomer. A crystal structure of the mitochondrial Hsp90, TRAP1, revealed that the catalytically active state is closed in a highly strained asymmetric conformation. This asymmetry, unobserved in other Hsp90 homologs, is due to buckling of one of the protomers and is most pronounced at the broadly conserved client-binding region. Here, we show that rather than being cooperative or independent, ATP hydrolysis on the two protomers is sequential and deterministic. Moreover, dimer asymmetry sets up differential hydrolysis rates for each protomer, such that the buckled conformation favors ATP hydrolysis. Remarkably, after the first hydrolysis, the dimer undergoes a flip in the asymmetry while remaining in a closed state for the second hydrolysis. From these results, we propose a model where direct coupling of ATP hydrolysis and conformational flipping rearranges client-binding sites, providing a paradigm of how energy from ATP hydrolysis can be used for client remodeling.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Howard Hughes Medical Institute, University of California, San Francisco, United States.