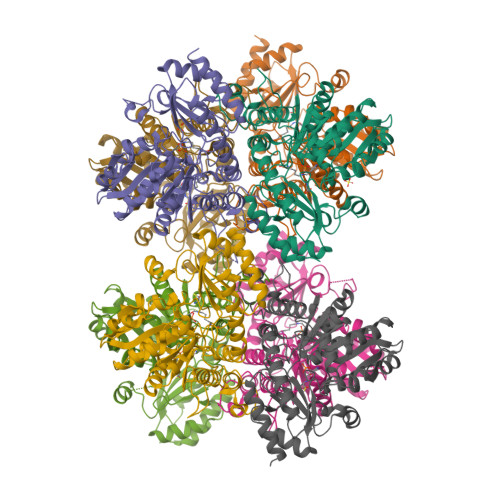
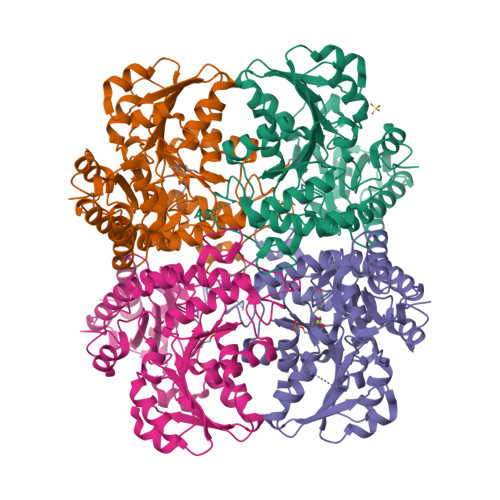
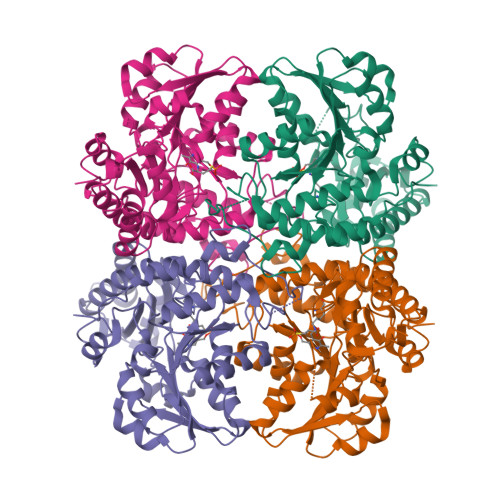
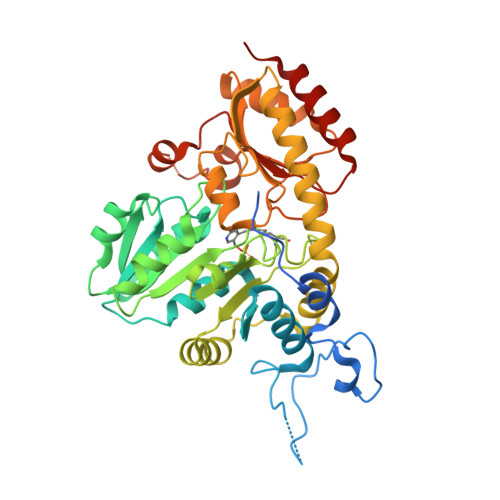
Structural Snapshots of an Engineered Cystathionine-gamma-lyase Reveal the Critical Role of Electrostatic Interactions in the Active Site.
Yan, W., Stone, E., Zhang, Y.J.(2017) Biochemistry 56: 876-885
- PubMed: 28106980
- DOI: https://doi.org/10.1021/acs.biochem.6b01172
- Primary Citation of Related Structures:
5TSU, 5TT2 - PubMed Abstract:
Enzyme therapeutics that can degrade l-methionine (l-Met) are of great interest as numerous malignancies are exquisitely sensitive to l-Met depletion. To exhaust the pool of methionine in human serum, we previously engineered an l-Met-degrading enzyme based on the human cystathionine-γ-lyase scaffold (hCGL-NLV) to circumvent immunogenicity and stability issues observed in the preclinical application of bacterially derived methionine-γ-lyases. To gain further insights into the structure-activity relationships governing the chemistry of the hCGL-NLV lead molecule, we undertook a biophysical characterization campaign that captured crystal structures (2.2 Å) of hCGL-NLV with distinct reaction intermediates, including internal aldimine, substrate-bound, gem-diamine, and external aldimine forms. Curiously, an alternate form of hCGL-NLV that crystallized under higher-salt conditions revealed a locally unfolded active site, correlating with inhibition of activity as a function of ionic strength. Subsequent mutational and kinetic experiments pinpointed that a salt bridge between the phosphate of the essential cofactor pyridoxal 5'-phosphate (PLP) and residue R62 plays an important role in catalyzing β- and γ-eliminations. Our study suggests that solvent ions such as NaCl disrupt electrostatic interactions between R62 and PLP, decreasing catalytic efficiency.
Organizational Affiliation:
Department of Molecular Biosciences and ‡Institute for Cellular and Molecular Biology, The University of Texas at Austin , Austin, Texas 78712, United States.