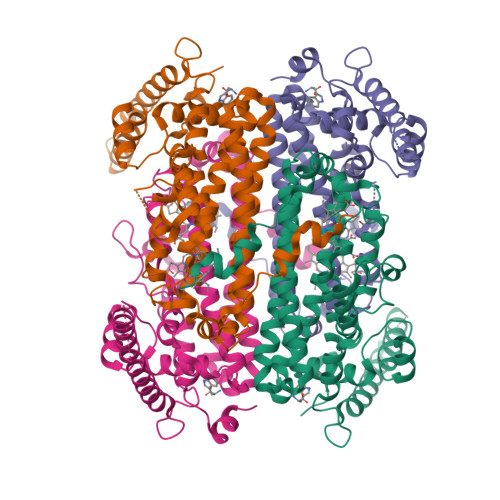
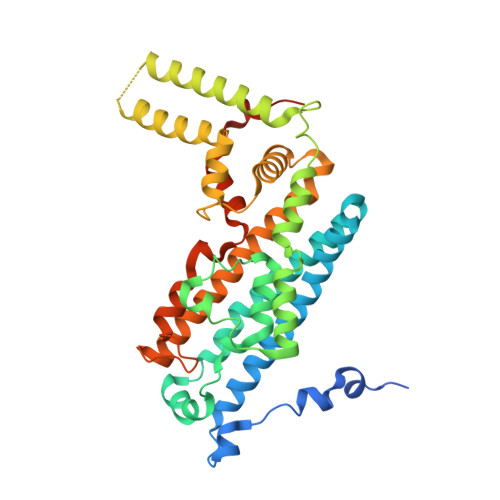
Molecular basis for catalysis and substrate-mediated cellular stabilization of human tryptophan 2,3-dioxygenase.
Lewis-Ballester, A., Forouhar, F., Kim, S.M., Lew, S., Wang, Y., Karkashon, S., Seetharaman, J., Batabyal, D., Chiang, B.Y., Hussain, M., Correia, M.A., Yeh, S.R., Tong, L.(2016) Sci Rep 6: 35169-35169
- PubMed: 27762317
- DOI: https://doi.org/10.1038/srep35169
- Primary Citation of Related Structures:
5TI9, 5TIA - PubMed Abstract:
Tryptophan 2,3-dioxygenase (TDO) and indoleamine 2,3-dioxygenase (IDO) play a central role in tryptophan metabolism and are involved in many cellular and disease processes. Here we report the crystal structure of human TDO (hTDO) in a ternary complex with the substrates L-Trp and O 2 and in a binary complex with the product N-formylkynurenine (NFK), defining for the first time the binding modes of both substrates and the product of this enzyme. The structure indicates that the dioxygenation reaction is initiated by a direct attack of O 2 on the C 2 atom of the L-Trp indole ring. The structure also reveals an exo binding site for L-Trp, located ~42 Å from the active site and formed by residues conserved among tryptophan-auxotrophic TDOs. Biochemical and cellular studies indicate that Trp binding at this exo site does not affect enzyme catalysis but instead it retards the degradation of hTDO through the ubiquitin-dependent proteasomal pathway. This exo site may therefore provide a novel L-Trp-mediated regulation mechanism for cellular degradation of hTDO, which may have important implications in human diseases.
Organizational Affiliation:
Department of Physiology and Biophysics Albert Einstein College of Medicine Bronx, NY 10461, USA.