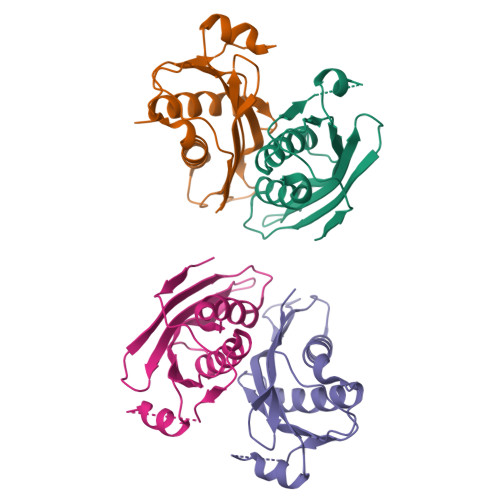
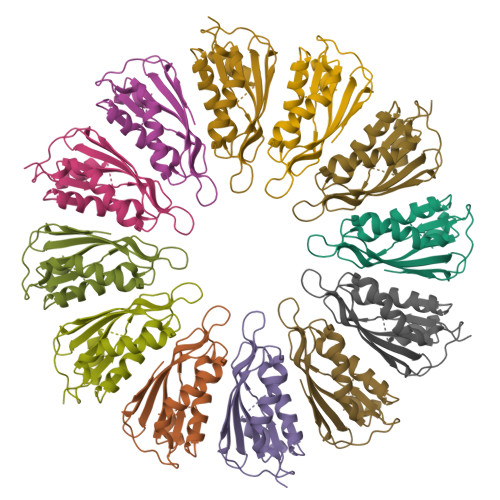
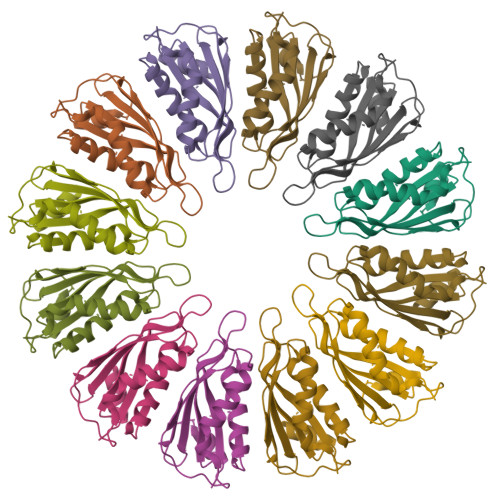
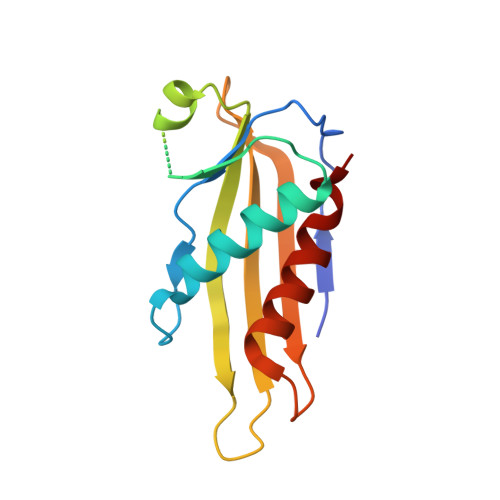
Oligomeric lipoprotein PelC guides Pel polysaccharide export across the outer membrane of Pseudomonas aeruginosa.
Marmont, L.S., Rich, J.D., Whitney, J.C., Whitfield, G.B., Almblad, H., Robinson, H., Parsek, M.R., Harrison, J.J., Howell, P.L.(2017) Proc Natl Acad Sci U S A 114: 2892-2897
- PubMed: 28242707
- DOI: https://doi.org/10.1073/pnas.1613606114
- Primary Citation of Related Structures:
5T0Z, 5T10, 5T11 - PubMed Abstract:
Secreted polysaccharides are important functional and structural components of bacterial biofilms. The opportunistic pathogen Pseudomonas aeruginosa produces the cationic exopolysaccharide Pel, which protects bacteria from aminoglycoside antibiotics and contributes to biofilm architecture through ionic interactions with extracellular DNA. A bioinformatics analysis of genome databases suggests that gene clusters for Pel biosynthesis are present in >125 bacterial species, yet little is known about how this biofilm exopolysaccharide is synthesized and exported from the cell. In this work, we characterize PelC, an outer membrane lipoprotein essential for Pel production. Crystal structures of PelC from Geobacter metallireducens and Paraburkholderia phytofirmans coupled with structure-guided disulfide cross-linking in P. aeruginosa suggest that PelC assembles into a 12- subunit ring-shaped oligomer. In this arrangement, an aromatic belt in proximity to its lipidation site positions the highly electronegative surface of PelC toward the periplasm. PelC is structurally similar to the Escherichia coli amyloid exporter CsgG; however, unlike CsgG, PelC does not possess membrane-spanning segments required for polymer export across the outer membrane. We show that the multidomain protein PelB with a predicted C-terminal β-barrel porin localizes to the outer membrane, and propose that PelC functions as an electronegative funnel to guide the positively charged Pel polysaccharide toward an exit channel formed by PelB. Together, our findings provide insight into the unique molecular architecture and export mechanism of the Pel apparatus, a widespread exopolysaccharide secretion system found in environmental and pathogenic bacteria.
Organizational Affiliation:
Program in Molecular Structure & Function, The Hospital for Sick Children, Toronto, ON, Canada M5G 0A4.