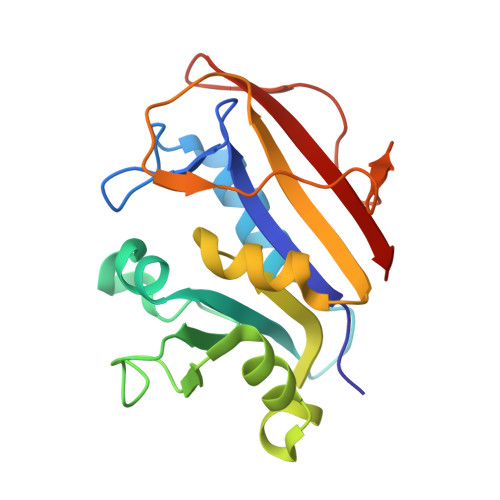
Crystal Structure of Dihydrofolate Reductase from Homo sapiens bound to NADP and SDDC Inhibitor SDDC-892
Mayclin, S.J., Fairman, J.W., Dranow, D.M., Conrady, D.G., Fox III, D., Lukacs, C.M., Lorimer, D.D., Horanyi, P.S., Edwards, T.E., Abendroth, J.To be published.