alpha-Helix or beta-Turn? An Investigation into N-Terminally Constrained Analogues of Glucagon-like Peptide 1 (GLP-1) and Exendin-4.
Oddo, A., Mortensen, S., Thogersen, H., De Maria, L., Hennen, S., McGuire, J.N., Kofoed, J., Linderoth, L., Reedtz-Runge, S.(2018) Biochemistry 57: 4148-4154
- PubMed: 29877701
- DOI: https://doi.org/10.1021/acs.biochem.8b00105
- Primary Citation of Related Structures:
5OTT, 5OTU, 5OTV, 5OTW, 5OTX - PubMed Abstract:
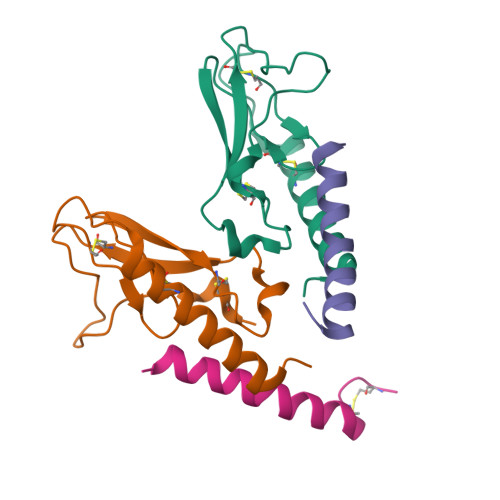
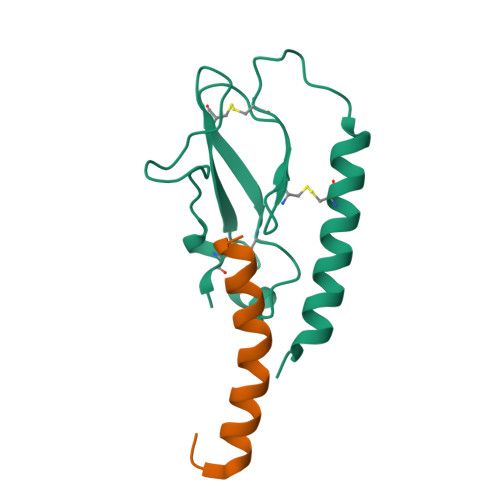
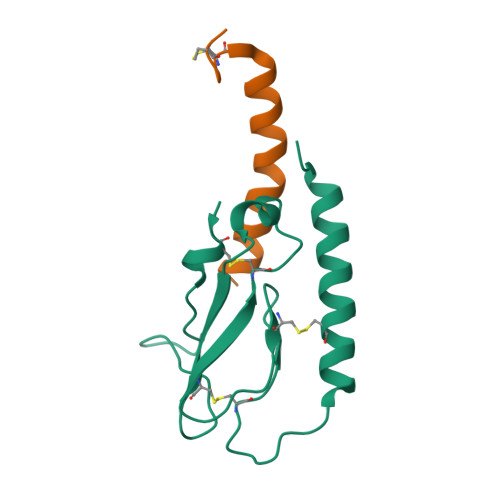
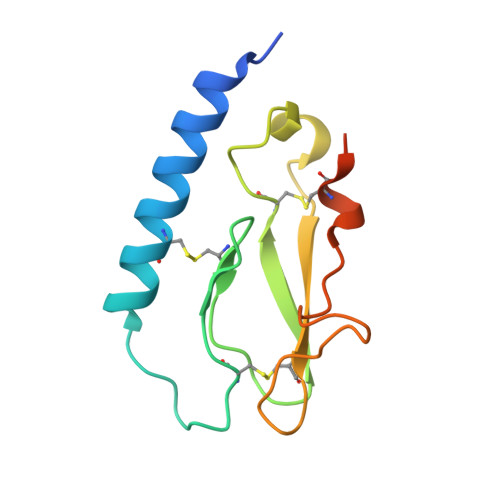
Peptide agonists acting on the glucagon-like peptide 1 receptor (GLP-1R) promote glucose-dependent insulin release and therefore represent important therapeutic agents for type 2 diabetes (T2D). Previous data indicated that an N-terminal type II β-turn motif might be an important feature for agonists acting on the GLP-1R. In contrast, recent publications reporting the structure of the full-length GLP-1R have shown the N-terminus of receptor-bound agonists in an α-helical conformation. To reconcile these conflicting results, we prepared N-terminally constrained analogues of glucagon-like peptide 1 (GLP-1) and exendin-4 and evaluated their receptor affinity and functionality in vitro; we then examined their crystal structures in complex with the extracellular domain of the GLP-1R and used molecular modeling and molecular dynamics simulations for further investigations. We report that the peptides' N-termini in all determined crystal structures adopted a type II β-turn conformation, but in vitro potency varied several thousand-fold across the series. Potency correlated better with α-helicity in our computational model, although we have found that the energy barrier between the two mentioned conformations is low in our most potent analogues and the flexibility of the N-terminus is highlighted by the dynamics simulations.
Organizational Affiliation:
Global Research , Novo Nordisk A/S , Novo Nordisk Park , 2760 Måløv , Denmark.