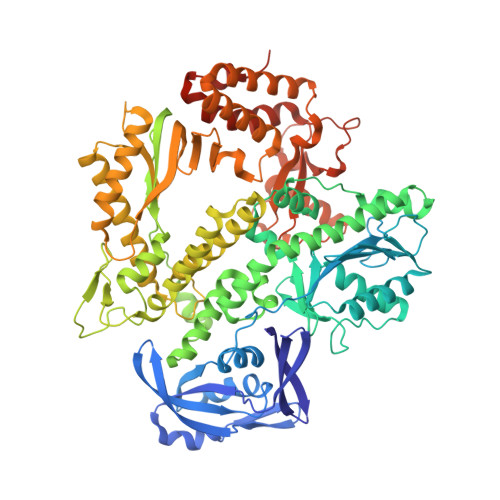
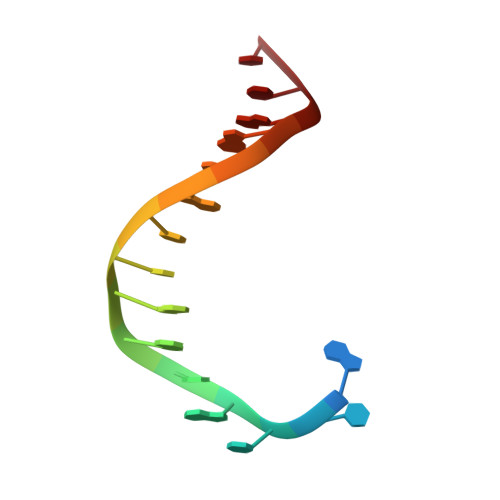
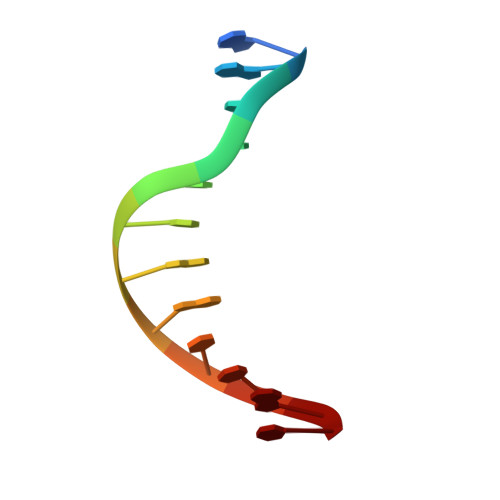
Crystal structures of ternary complexes of archaeal B-family DNA polymerases.
Kropp, H.M., Betz, K., Wirth, J., Diederichs, K., Marx, A.(2017) PLoS One 12: e0188005-e0188005
- PubMed: 29211756
- DOI: https://doi.org/10.1371/journal.pone.0188005
- Primary Citation of Related Structures:
5OMF, 5OMQ, 5OMV - PubMed Abstract:
Archaeal B-family polymerases drive biotechnology by accepting a wide substrate range of chemically modified nucleotides. By now no structural data for archaeal B-family DNA polymerases in a closed, ternary complex are available, which would be the basis for developing next generation nucleotides. We present the ternary crystal structures of KOD and 9°N DNA polymerases complexed with DNA and the incoming dATP. The structures reveal a third metal ion in the active site, which was so far only observed for the eukaryotic B-family DNA polymerase δ and no other B-family DNA polymerase. The structures reveal a wide inner channel and numerous interactions with the template strand that provide space for modifications within the enzyme and may account for the high processivity, respectively. The crystal structures provide insights into the superiority over other DNA polymerases concerning the acceptance of modified nucleotides.
- Konstanz Research School Chemical Biology, University of Konstanz, Baden-Württemberg, Konstanz, Germany.
Organizational Affiliation: