Fluorescent Retinoic Acid Analogues as Probes for Biochemical and Intracellular Characterization of Retinoid Signaling Pathways.
Chisholm, D.R., Tomlinson, C.W.E., Zhou, G.L., Holden, C., Affleck, V., Lamb, R., Newling, K., Ashton, P., Valentine, R., Redfern, C., Erostyak, J., Makkai, G., Ambler, C.A., Whiting, A., Pohl, E.(2019) ACS Chem Biol 14: 369-377
- PubMed: 30707838
- DOI: https://doi.org/10.1021/acschembio.8b00916
- Primary Citation of Related Structures:
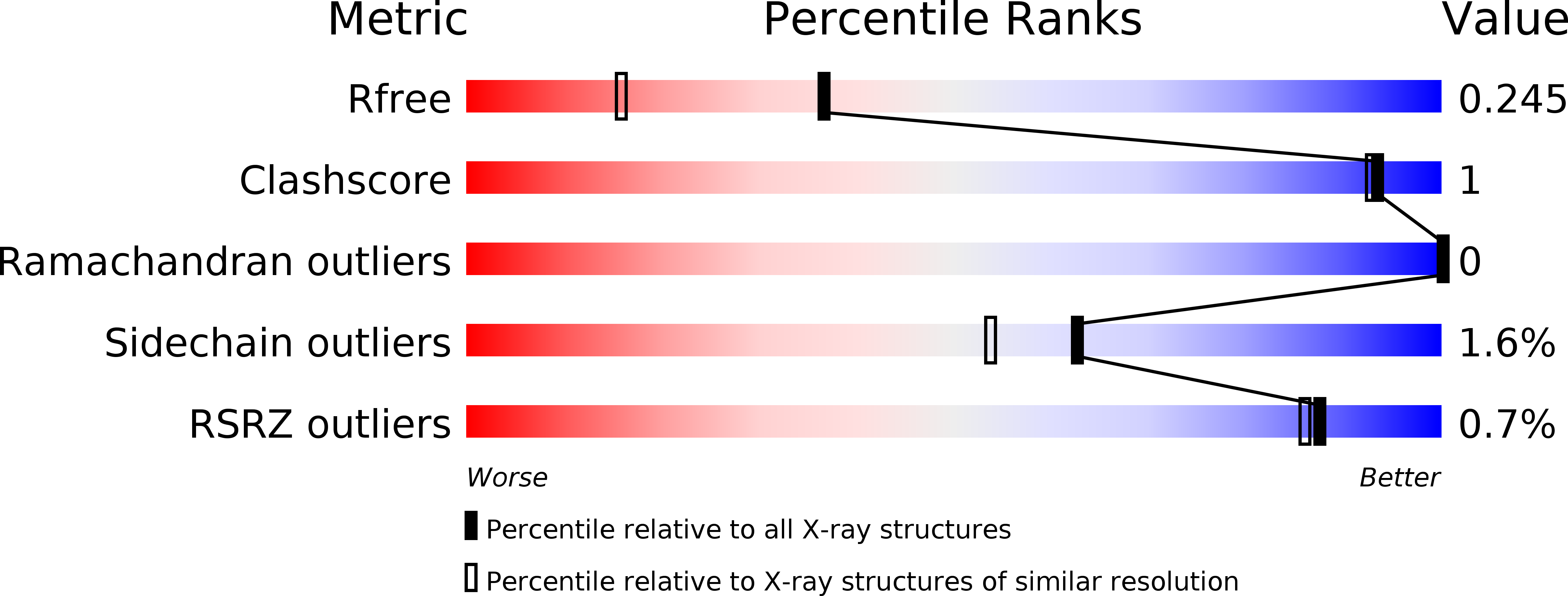
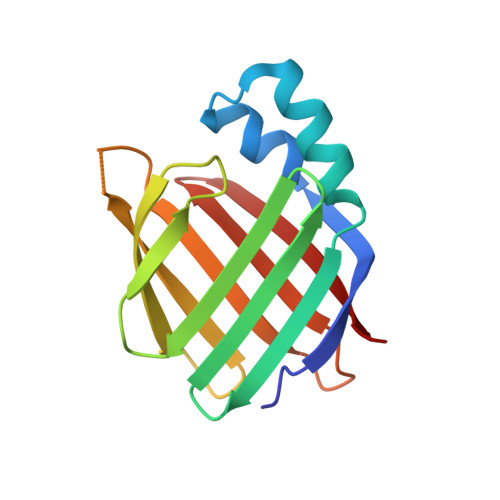
5OGB - PubMed Abstract:
Retinoids, such as all- trans-retinoic acid (ATRA), are endogenous signaling molecules derived from vitamin A that influence a variety of cellular processes through mediation of transcription events in the cell nucleus. Because of these wide-ranging and powerful biological activities, retinoids have emerged as therapeutic candidates of enormous potential. However, their use has been limited, to date, due to a lack of understanding of the complex and intricate signaling pathways that they control. We have designed and synthesized a family of synthetic retinoids that exhibit strong, intrinsic, solvatochromatic fluorescence as multifunctional tools to interrogate these important biological activities. We utilized the unique photophysical characteristics of these fluorescent retinoids to develop a novel in vitro fluorometric binding assay to characterize and quantify their binding to their cellular targets, including cellular retinoid binding protein II (CRABPII). The dihydroquinoline retinoid, DC360, exhibited particularly strong binding ( K d = 34.0 ± 2.5 nM), and we further used X-ray crystallography to determine the structure of the DC360-CRABPII complex to 1.8 Å, which showed that DC360 occupies the known hydrophobic retinoid binding pocket. Finally, we used confocal fluorescence microscopy to image the cellular behavior of the compounds in cultured human epithelial cells, highlighting a fascinating nuclear localization, and used RNA sequencing to confirm that the compounds regulate cellular processes similar to those of ATRA. We anticipate that the unique properties of these fluorescent retinoids can now be used to cast new light on the vital and highly complex retinoid signaling pathway.
Organizational Affiliation:
Department of Chemistry , Durham University , South Road , Durham DH1 3LE , U.K.