Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Lu, K.Y., Chen, W.F., Rety, S., Liu, N.N., Wu, W.Q., Dai, Y.X., Li, D., Ma, H.Y., Dou, S.X., Xi, X.G.(2018) Nucleic Acids Res 46: 1486-1500
- PubMed: 29202194
- DOI: https://doi.org/10.1093/nar/gkx1217
- Primary Citation of Related Structures:
5O6B, 5O6D, 5O6E - PubMed Abstract:
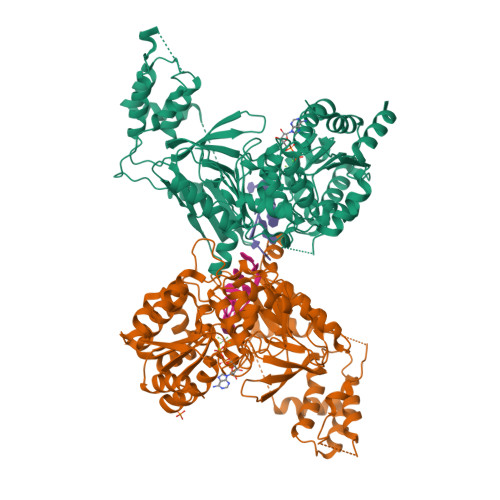
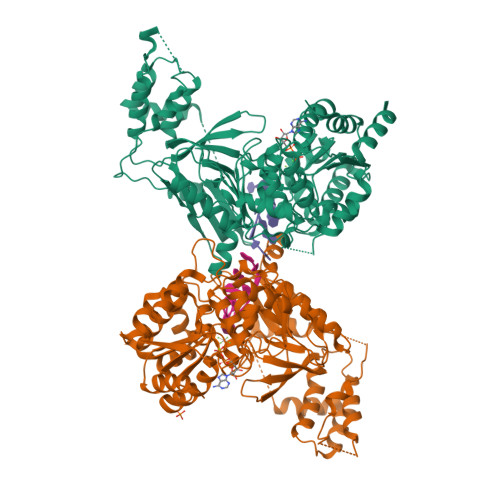
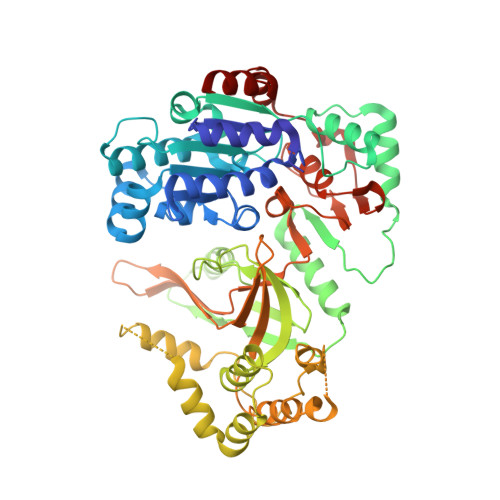
The Saccharomyces cerevisiae Pif1 protein (ScPif1p) is the prototypical member of the Pif1 family of DNA helicases. ScPif1p is involved in the maintenance of mitochondrial, ribosomal and telomeric DNA and suppresses genome instability at G-quadruplex motifs. Here, we report the crystal structures of a truncated ScPif1p (ScPif1p237-780) in complex with different ssDNAs. Our results have revealed that a yeast-specific insertion domain protruding from the 2B domain folds as a bundle bearing an α-helix, α16. The α16 helix regulates the helicase activities of ScPif1p through interactions with the previously identified loop3. Furthermore, a biologically relevant dimeric structure has been identified, which can be further specifically stabilized by G-quadruplex DNA. Basing on structural analyses and mutational studies with DNA binding and unwinding assays, a potential G-quadruplex DNA binding site in ScPif1p monomers is suggested. Our results also show that ScPif1p uses the Q-motif to preferentially hydrolyze ATP, and a G-rich tract is preferentially recognized by more residues, consistent with previous biochemical observations. These findings provide a structural and mechanistic basis for understanding the multifunctional ScPif1p.
Organizational Affiliation:
College of Life Sciences, Northwest A&F University, Yangling, Shaanxi 712100, China.