A study of the reactivity of S(VI)-F containing warheads with nucleophilic amino-acid side chains under physiological conditions.
Mukherjee, H., Debreczeni, J., Breed, J., Tentarelli, S., Aquila, B., Dowling, J.E., Whitty, A., Grimster, N.P.(2017) Org Biomol Chem 15: 9685-9695
- PubMed: 29119993
- DOI: https://doi.org/10.1039/c7ob02028g
- Primary Citation of Related Structures:
5O49, 5O4A - PubMed Abstract:
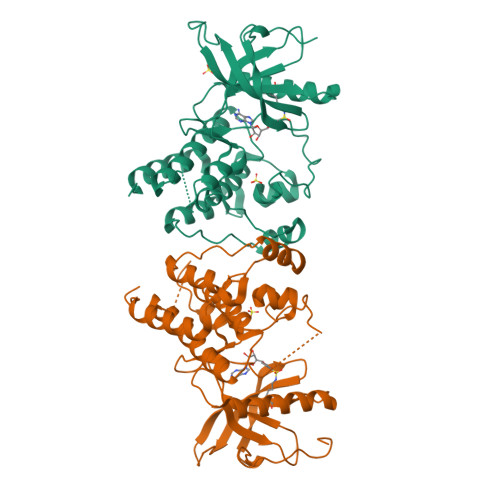
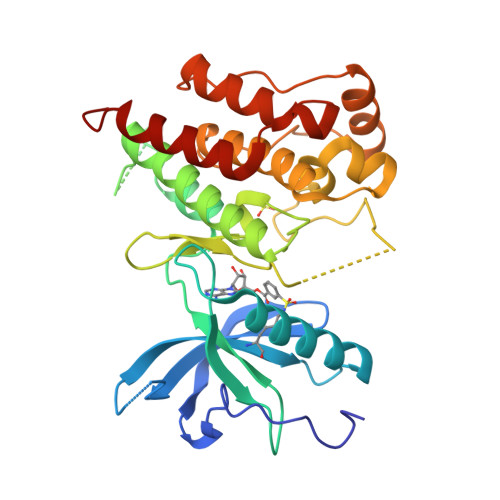
Sulfonyl fluorides (SFs) have recently emerged as a promising warhead for the targeted covalent modification of proteins. Despite numerous examples of the successful deployment of SFs as covalent probe compounds, a detailed exploration of the factors influencing the stability and reactivity of SFs has not yet appeared. In this work we present an extensive study on the influence of steric and electronic factors on the reactivity and stability of the SF and related S VI -F groups. While SFs react rapidly with N-acetylcysteine, the resulting adducts were found to be unstable, rendering SFs inappropriate for the durable covalent inhibition of cysteine residues. In contrast, SFs afforded stable adducts with both N-acetyltyrosine and N-acetyllysine; furthermore, we show that the reactivity of arylsulfonyl fluorides towards these nucleophilic amino acids can be predictably modulated by adjusting the electronic properties of the warhead. These trends were largely conserved when the covalent reaction occurred within a protein binding pocket. We have also obtained a crystal structure depicting covalent modification of the catalytic lysine of a tyrosine kinase (FGFR1) by the ATP analog 5'-O-3-((fluorosulfonyl)benzoyl)adenosine (m-FSBA). Highly reactive warheads were demonstrated to be unstable with respect to hydrolysis in buffered aqueous solutions, indicating that warhead reactivity must be carefully tuned to provide optimal rates of protein modification. Our results demonstrate that the reactivity of SFs complements that of more commonly studied acrylamides, and we hope that this work spurs the rational design of novel SF-containing covalent probe compounds and inhibitors, particularly in cases where a suitably positioned cysteine residue is not present.
Organizational Affiliation:
IMED Oncology, AstraZeneca, Waltham, Massachusetts 02451, USA. neil.grimster@astrazeneca.com.