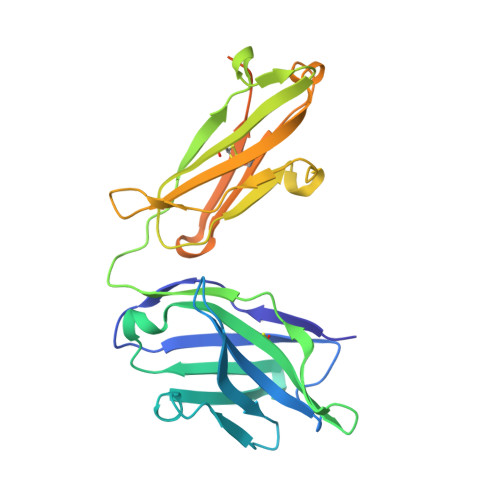
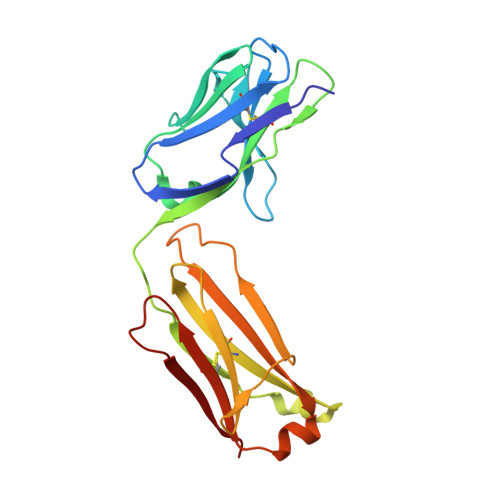
Structures of NHBA elucidate a broadly conserved epitope identified by a vaccine induced antibody.
Maritan, M., Veggi, D., Cozzi, R., Dello Iacono, L., Bartolini, E., Lo Surdo, P., Maruggi, G., Spraggon, G., Bottomley, M.J., Malito, E.(2018) PLoS One 13: e0201922-e0201922
- PubMed: 30133484
- DOI: https://doi.org/10.1371/journal.pone.0201922
- Primary Citation of Related Structures:
5NYX, 5O1R, 6CUJ - PubMed Abstract:
Neisserial heparin binding antigen (NHBA) is one of three main recombinant protein antigens in 4CMenB, a vaccine for the prevention of invasive meningococcal disease caused by Neisseria meningitidis serogroup B. NHBA is a surface-exposed lipoprotein composed of a predicted disordered N-terminal region, an arginine-rich region that binds heparin, and a C-terminal domain that folds as an anti-parallel β-barrel and that upon release after cleavage by human proteases alters endothelial permeability. NHBA induces bactericidal antibodies in humans, and NHBA-specific antibodies elicited by the 4CMenB vaccine contribute to serum bactericidal activity, the correlate of protection. To better understand the structural bases of the human antibody response to 4CMenB vaccination and to inform antigen design, we used X-ray crystallography to elucidate the structures of two C-terminal fragments of NHBA, either alone or in complex with the Fab derived from the vaccine-elicited human monoclonal antibody 5H2, and the structure of the unbound Fab 5H2. The structures reveal details on the interaction between an N-terminal β-hairpin fragment and the β-barrel, and explain how NHBA is capable of generating cross-reactive antibodies through an extensive conserved conformational epitope that covers the entire C-terminal face of the β-barrel. By providing new structural information on a vaccine antigen and on the human immune response to vaccination, these results deepen our molecular understanding of 4CMenB, and might also aid future vaccine design projects.
Organizational Affiliation:
GSK, Siena, Italy.