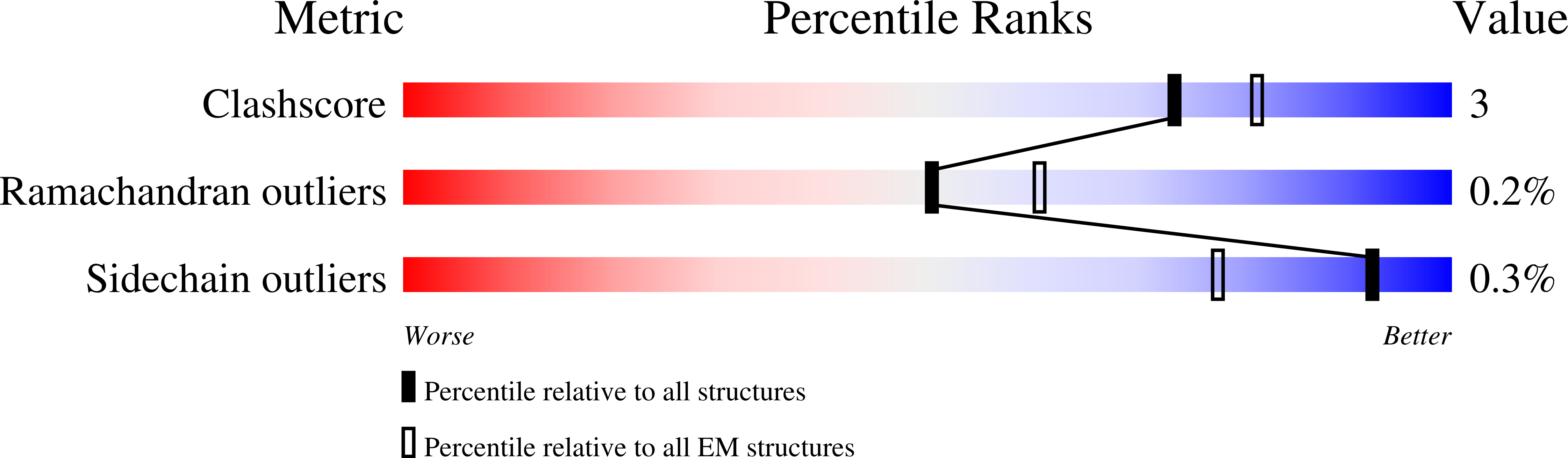
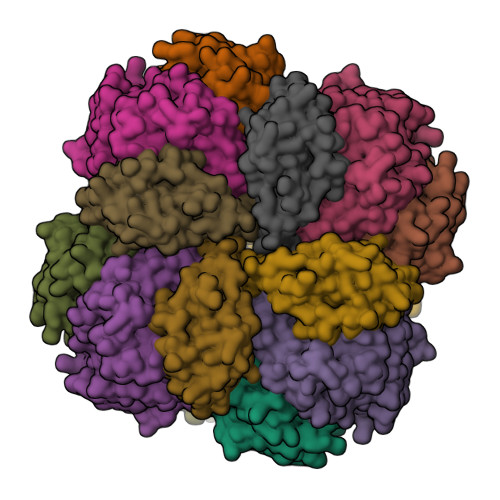
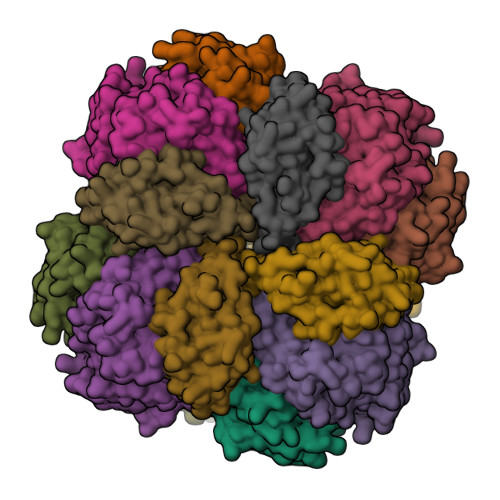
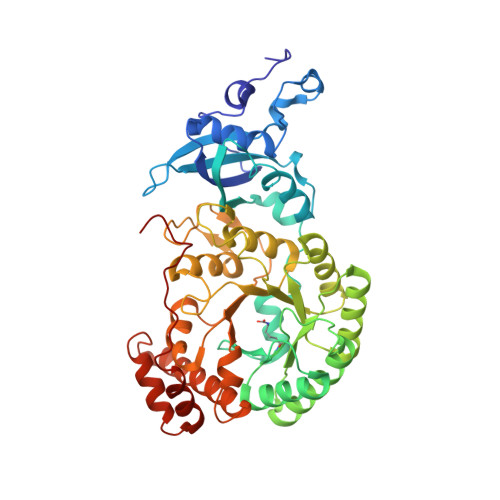
Mechanism of Enzyme Repair by the AAA(+) Chaperone Rubisco Activase.
Bhat, J.Y., Milicic, G., Thieulin-Pardo, G., Bracher, A., Maxwell, A., Ciniawsky, S., Mueller-Cajar, O., Engen, J.R., Hartl, F.U., Wendler, P., Hayer-Hartl, M.(2017) Mol Cell 67: 744-756.e6
- PubMed: 28803776
- DOI: https://doi.org/10.1016/j.molcel.2017.07.004
- Primary Citation of Related Structures:
5NV3 - PubMed Abstract:
How AAA+ chaperones conformationally remodel specific target proteins in an ATP-dependent manner is not well understood. Here, we investigated the mechanism of the AAA+ protein Rubisco activase (Rca) in metabolic repair of the photosynthetic enzyme Rubisco, a complex of eight large (RbcL) and eight small (RbcS) subunits containing eight catalytic sites. Rubisco is prone to inhibition by tight-binding sugar phosphates, whose removal is catalyzed by Rca. We engineered a stable Rca hexamer ring and analyzed its functional interaction with Rubisco. Hydrogen/deuterium exchange and chemical crosslinking showed that Rca structurally destabilizes elements of the Rubisco active site with remarkable selectivity. Cryo-electron microscopy revealed that Rca docks onto Rubisco over one active site at a time, positioning the C-terminal strand of RbcL, which stabilizes the catalytic center, for access to the Rca hexamer pore. The pulling force of Rca is fine-tuned to avoid global destabilization and allow for precise enzyme repair.
Organizational Affiliation:
Department of Cellular Biochemistry, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany.