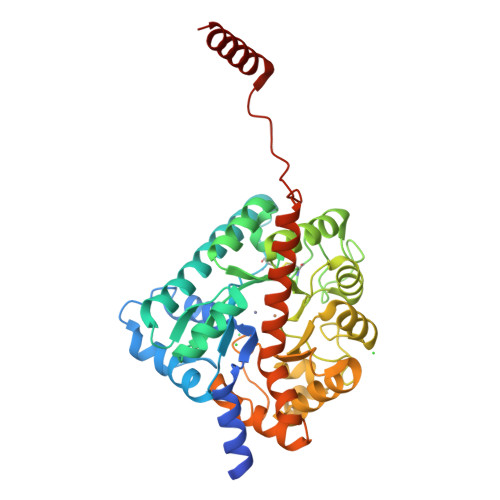
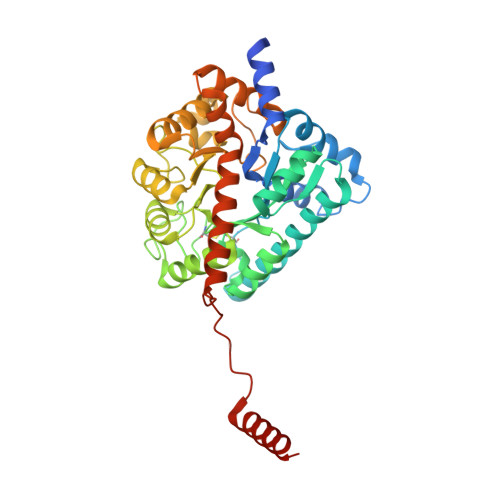
Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
Marion, A., Groll, M., Scharf, D.H., Scherlach, K., Glaser, M., Sievers, H., Schuster, M., Hertweck, C., Brakhage, A.A., Antes, I., Huber, E.M.(2017) ACS Chem Biol 12: 1874-1882
- PubMed: 28525266
- DOI: https://doi.org/10.1021/acschembio.6b00847
- Primary Citation of Related Structures:
5LWZ, 5LX0, 5LX1, 5LX4, 5LX7, 5NRT, 5NRU, 5NRX, 5NRY, 5NRZ, 5NS1, 5NS2, 5NS5 - PubMed Abstract:
The formation of glutathione (GSH) conjugates, best known from the detoxification of xenobiotics, is a widespread strategy to incorporate sulfur into biomolecules. The biosynthesis of gliotoxin, a virulence factor of the human pathogenic fungus Aspergillus fumigatus, involves attachment of two GSH molecules and their sequential decomposition to yield two reactive thiol groups. The degradation of the GSH moieties requires the activity of the Cys-Gly carboxypeptidase GliJ, for which we describe the X-ray structure here. The enzyme forms a homodimer with each monomer comprising one active site. Two metal ions are present per proteolytic center, thus assigning GliJ to the diverse family of dinuclear metallohydrolases. Depending on availability, Zn 2+ , Fe 2+ , Fe 3+ , Mn 2+ , Cu 2+ , Co 2+ , or Ni 2+ ions are accepted as cofactors. Despite this high metal promiscuity, a preference for zinc versus iron and manganese was noted. Mutagenesis experiments revealed details of metal coordination, and molecular modeling delivered insights into substrate recognition and processing by GliJ. The latter results suggest a reaction mechanism in which the two scissile peptide bonds of one gliotoxin precursor molecule are hydrolyzed sequentially and in a given order.
Organizational Affiliation:
Center for Integrated Protein Science Munich at the Department of Biosciences, Technische Universität München , Emil-Erlenmeyer-Forum 8, D-85354 Freising, Germany.