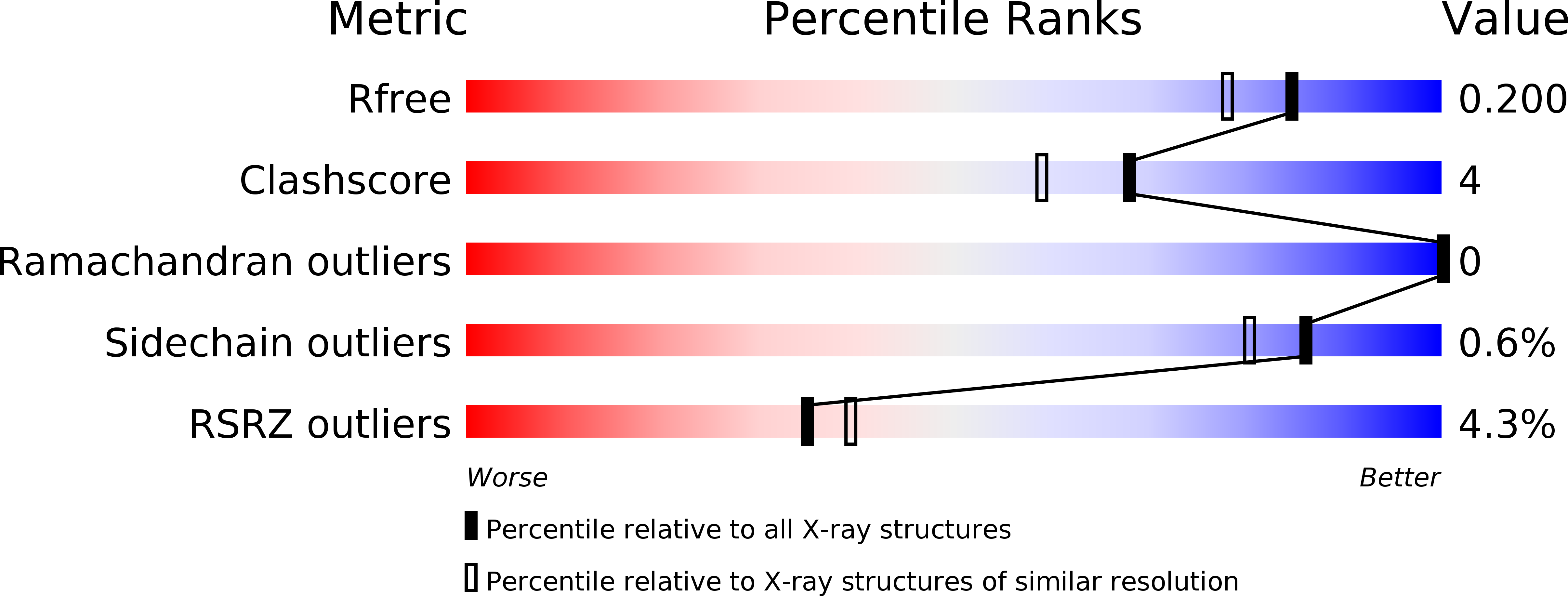
Nonribosomal biosynthesis of backbone-modified peptides.
Niquille, D.L., Hansen, D.A., Mori, T., Fercher, D., Kries, H., Hilvert, D.(2018) Nat Chem 10: 282-287
- PubMed: 29461527
- DOI: https://doi.org/10.1038/nchem.2891
- Primary Citation of Related Structures:
5N81, 5N82 - PubMed Abstract:
Biosynthetic modification of nonribosomal peptide backbones represents a potentially powerful strategy to modulate the structure and properties of an important class of therapeutics. Using a high-throughput assay for catalytic activity, we show here that an L-Phe-specific module of an archetypal nonribosomal peptide synthetase can be reprogrammed to accept and process the backbone-modified amino acid (S)-β-Phe with near-native specificity and efficiency. A co-crystal structure with a non-hydrolysable aminoacyl-AMP analogue reveals the origins of the 40,000-fold α/β-specificity switch, illuminating subtle but precise remodelling of the active site. When the engineered catalyst was paired with downstream module(s), (S)-β-Phe-containing peptides were produced at preparative scale in vitro (~1 mmol) and high titres in vivo (~100 mg l -1 ), highlighting the potential of biosynthetic pathway engineering for the construction of novel nonribosomal β-frameworks.
Organizational Affiliation:
Laboratory of Organic Chemistry, ETH Zurich, 8093 Zurich, Switzerland.