Structures and enzymatic mechanisms of phycobiliprotein lyases CpcE/F and PecE/F.
Zhao, C., Hoppner, A., Xu, Q.Z., Gartner, W., Scheer, H., Zhou, M., Zhao, K.H.(2017) Proc Natl Acad Sci U S A 114: 13170-13175
- PubMed: 29180420
- DOI: https://doi.org/10.1073/pnas.1715495114
- Primary Citation of Related Structures:
5N3U - PubMed Abstract:
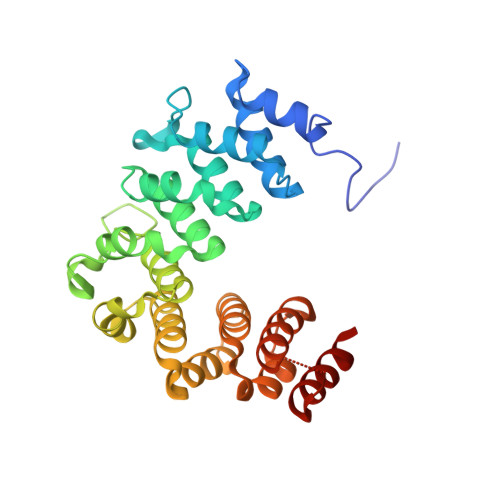
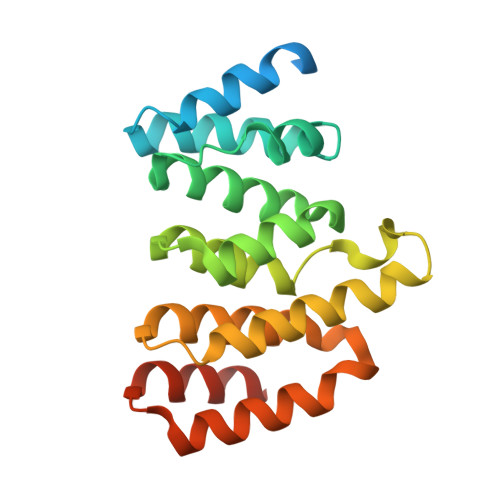
The light-harvesting phycobilisome in cyanobacteria and red algae requires the lyase-catalyzed chromophorylation of phycobiliproteins. There are three functionally distinct lyase families known. The heterodimeric E/F type is specific for attaching bilins covalently to α-subunits of phycocyanins and phycoerythrins. Unlike other lyases, the lyase also has chromophore-detaching activity. A subclass of the E/F-type lyases is, furthermore, capable of chemically modifying the chromophore. Although these enzymes were characterized >25 y ago, their structures remained unknown. We determined the crystal structure of the heterodimer of CpcE/F from Nostoc sp. PCC7120 at 1.89-Å resolution. Both subunits are twisted, crescent-shaped α-solenoid structures. CpcE has 15 and CpcF 10 helices. The inner (concave) layer of CpcE (helices h2, 4, 6, 8, 10, 12, and 14) and the outer (convex) layer of CpcF (h16, 18, 20, 22, and 24) form a cavity into which the phycocyanobilin chromophore can be modeled. This location of the chromophore is supported by mutations at the interface between the subunits and within the cavity. The structure of a structurally related, isomerizing lyase, PecE/F, that converts phycocyanobilin into phycoviolobilin, was modeled using the CpcE/F structure as template. A H 87 C 88 motif critical for the isomerase activity of PecE/F is located at the loop between h20 and h21, supporting the proposal that the nucleophilic addition of Cys-88 to C10 of phycocyanobilin induces the isomerization of phycocyanobilin into phycoviolobilin. Also, the structure of NblB, involved in phycobilisome degradation could be modeled using CpcE as template. Combined with CpcF, NblB shows a low chromophore-detaching activity.
Organizational Affiliation:
State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan 430070, P.R. China.