Preclinical pharmacology of AZD9977: A novel mineralocorticoid receptor modulator separating organ protection from effects on electrolyte excretion.
Bamberg, K., Johansson, U., Edman, K., William-Olsson, L., Myhre, S., Gunnarsson, A., Geschwindner, S., Aagaard, A., Bjornson Granqvist, A., Jaisser, F., Huang, Y., Granberg, K.L., Jansson-Lofmark, R., Hartleib-Geschwindner, J.(2018) PLoS One 13: e0193380-e0193380
- PubMed: 29474466
- DOI: https://doi.org/10.1371/journal.pone.0193380
- Primary Citation of Related Structures:
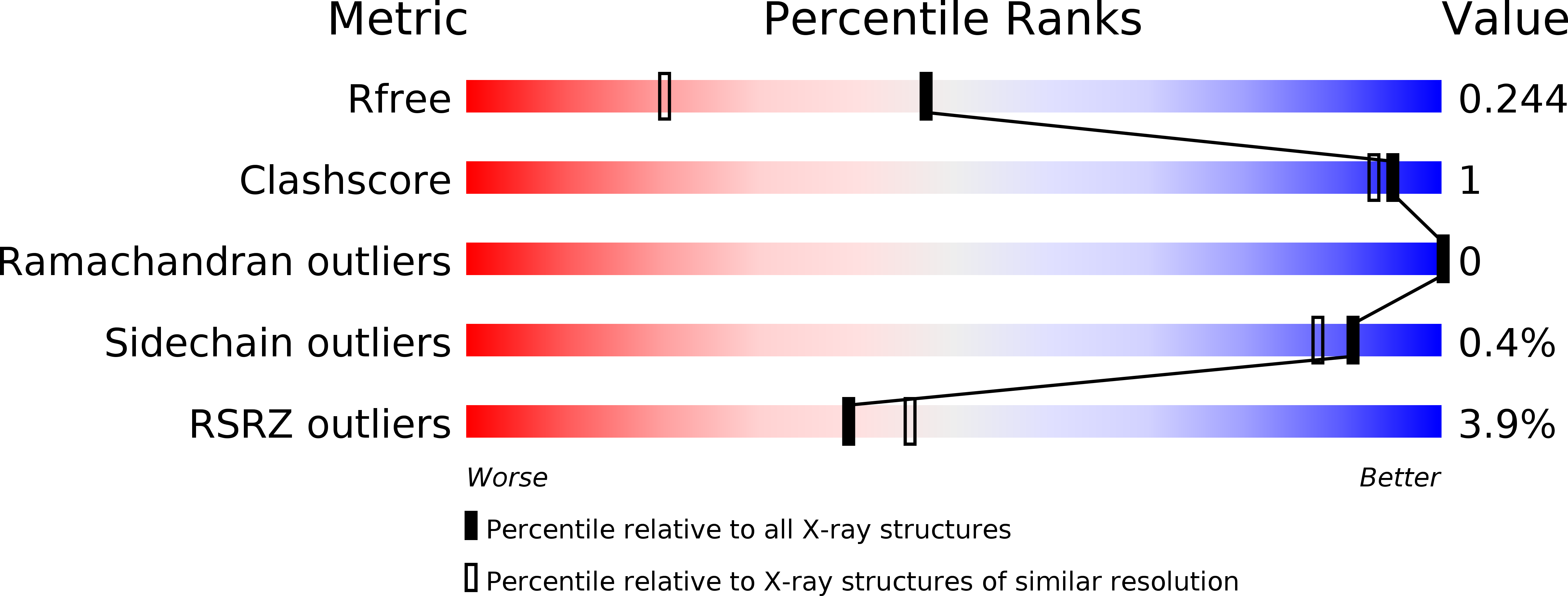
5MWP, 5MWY - PubMed Abstract:
Excess mineralocorticoid receptor (MR) activation promotes target organ dysfunction, vascular injury and fibrosis. MR antagonists like eplerenone are used for treating heart failure, but their use is limited due to the compound class-inherent hyperkalemia risk. Here we present evidence that AZD9977, a first-in-class MR modulator shows cardio-renal protection despite a mechanism-based reduced liability to cause hyperkalemia. AZD9977 in vitro potency and binding mode to MR were characterized using reporter gene, binding, cofactor recruitment assays and X-ray crystallopgraphy. Organ protection was studied in uni-nephrectomised db/db mice and uni-nephrectomised rats administered aldosterone and high salt. Acute effects of single compound doses on urinary electrolyte excretion were tested in rats on a low salt diet. AZD9977 and eplerenone showed similar human MR in vitro potencies. Unlike eplerenone, AZD9977 is a partial MR antagonist due to its unique interaction pattern with MR, which results in a distinct recruitment of co-factor peptides when compared to eplerenone. AZD9977 dose dependently reduced albuminuria and improved kidney histopathology similar to eplerenone in db/db uni-nephrectomised mice and uni-nephrectomised rats. In acute testing, AZD9977 did not affect urinary Na+/K+ ratio, while eplerenone increased the Na+/K+ ratio dose dependently. AZD9977 is a selective MR modulator, retaining organ protection without acute effect on urinary electrolyte excretion. This predicts a reduced hyperkalemia risk and AZD9977 therefore has the potential to deliver a safe, efficacious treatment to patients prone to hyperkalemia.
Organizational Affiliation:
Cardiovascular, Renal and Metabolic Diseases, Innovative Medicines and Early Development Biotech Unit, AstraZeneca, Gothenburg, Sweden.