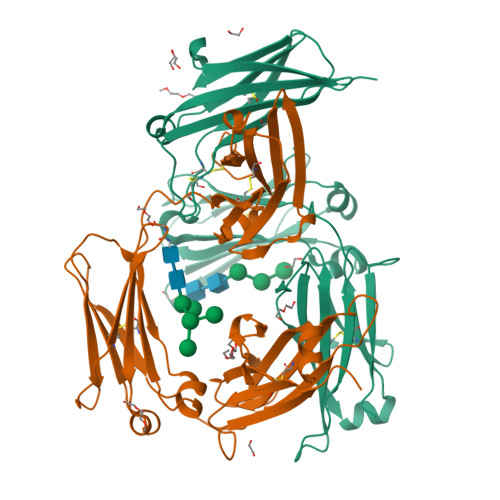
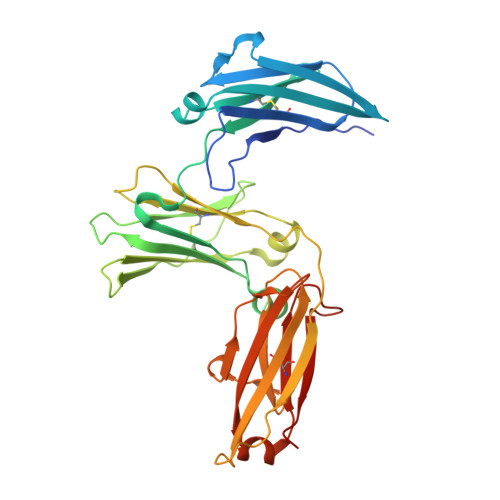
Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Dore, K.A., Davies, A.M., Drinkwater, N., Beavil, A.J., McDonnell, J.M., Sutton, B.J.(2017) Biochim Biophys Acta 1865: 1336-1347
- PubMed: 28844738
- DOI: https://doi.org/10.1016/j.bbapap.2017.08.005
- Primary Citation of Related Structures:
5MOI, 5MOJ, 5MOK, 5MOL - PubMed Abstract:
Immunoglobulin E (IgE) is the antibody that plays a central role in the mechanisms of allergic diseases such as asthma. Interactions with its receptors, FcεRI on mast cells and CD23 on B cells, are mediated by the Fc region, a dimer of the Cε2, Cε3 and Cε4 domains. A sub-fragment lacking the Cε2 domains, Fcε3-4, also binds to both receptors, although receptor binding almost exclusively involves the Cε3 domains. This domain also contains the N-linked glycosylation site conserved in other isotypes. We report here the crystal structures of IgE-Fc and Fcε3-4 at the highest resolutions yet determined, 1.75Å and 2.0Å respectively, revealing unprecedented detail regarding the carbohydrate and its interactions with protein domains. Analysis of the crystallographic B-factors of these, together with all earlier IgE-Fc and Fcε3-4 structures, shows that the Cε3 domains exhibit the greatest intrinsic flexibility and quaternary structural variation within IgE-Fc. Intriguingly, both well-ordered carbohydrate and disordered polypeptide can be seen within the same Cε3 domain. A simplified method for comparing the quaternary structures of the Cε3 domains in free and receptor-bound IgE-Fc structures is presented, which clearly delineates the FcεRI and CD23 bound states. Importantly, differential scanning fluorimetric analysis of IgE-Fc and Fcε3-4 identifies Cε3 as the domain most susceptible to thermally-induced unfolding, and responsible for the characteristically low melting temperature of IgE.
Organizational Affiliation:
King's College London, Randall Division of Cell and Molecular Biophysics, New Hunt's House, London SE1 1UL, United Kingdom; Medical Research Council & Asthma UK Centre in Allergic Mechanisms of Asthma, London, United Kingdom.