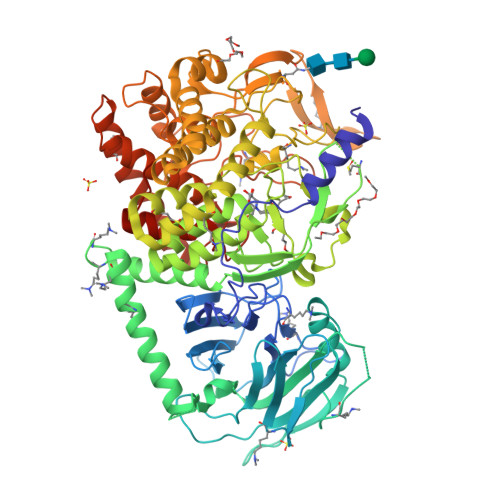
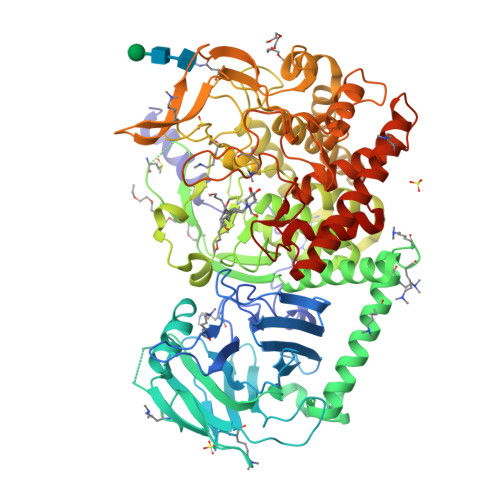
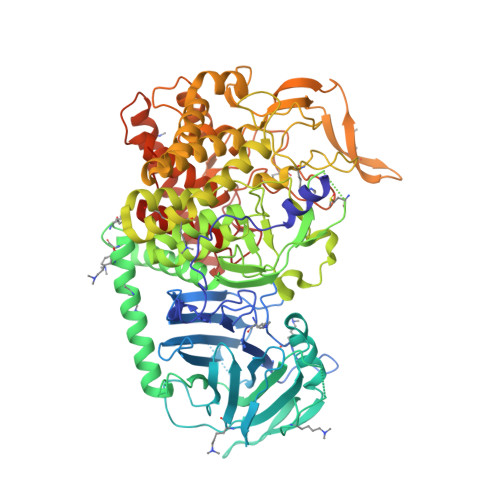
Targeting Endoplasmic Reticulum alpha-Glucosidase I with a Single-Dose Iminosugar Treatment Protects against Lethal Influenza and Dengue Virus Infections.
Warfield, K.L., Alonzi, D.S., Hill, J.C., Caputo, A.T., Roversi, P., Kiappes, J.L., Sheets, N., Duchars, M., Dwek, R.A., Biggins, J., Barnard, D., Shresta, S., Treston, A.M., Zitzmann, N.(2020) J Med Chem
- PubMed: 32227946
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00067
- Primary Citation of Related Structures:
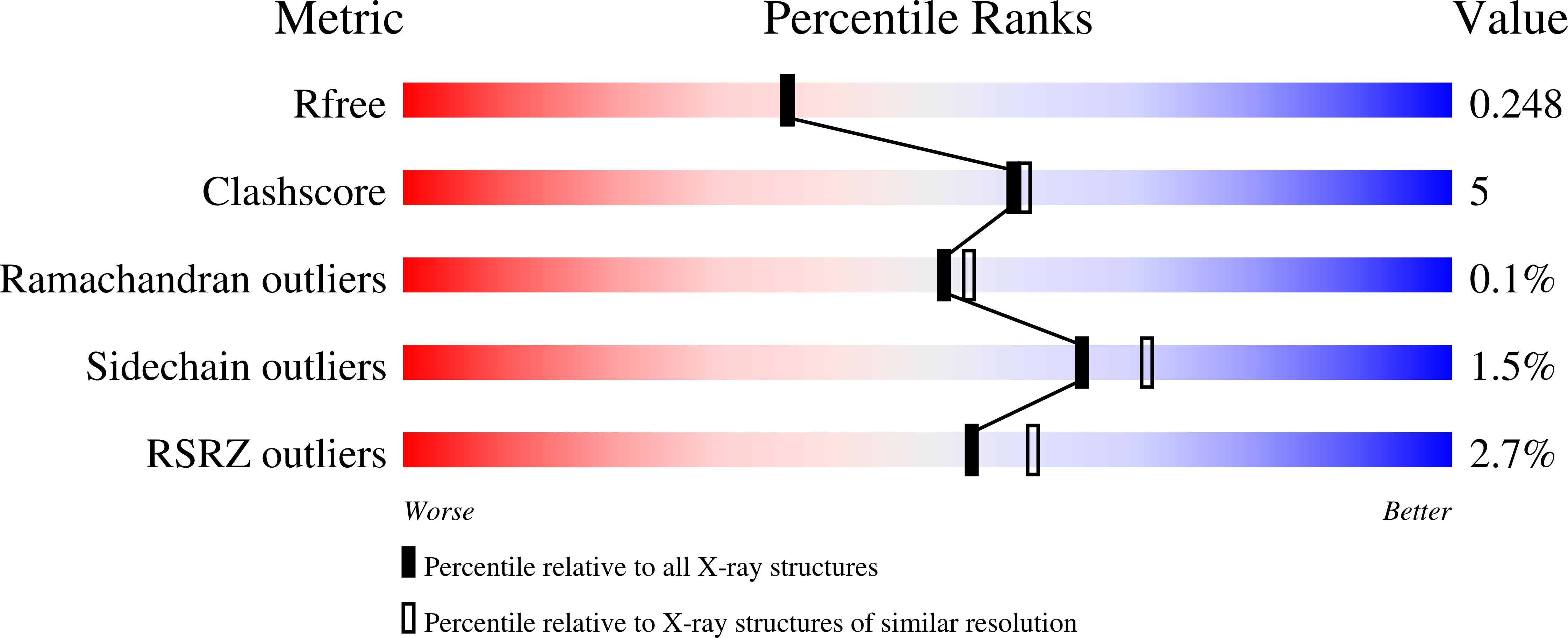
5MHF - PubMed Abstract:
Influenza and dengue viruses present a growing global threat to public health. Both viruses depend on the host endoplasmic reticulum (ER) glycoprotein folding pathway. In 2014, Sadat et al. reported two siblings with a rare genetic defect in ER α-glucosidase I (ER Glu I) who showed resistance to viral infections, identifying ER Glu I as a key antiviral target. Here, we show that a single dose of UV-4B (the hydrochloride salt form of N -(9'-methoxynonyl)-1-deoxynojirimycin; MON-DNJ) capable of inhibiting Glu I in vivo is sufficient to prevent death in mice infected with lethal viral doses, even when treatment is started as late as 48 h post infection. The first crystal structure of mammalian ER Glu I will constitute the basis for the development of potent and selective inhibitors. Targeting ER Glu I with UV-4B-derived compounds may alter treatment paradigms for acute viral disease through development of a single-dose therapeutic regime.
Organizational Affiliation:
Emergent BioSolutions, Gaithersburg, Maryland 20879, United States.