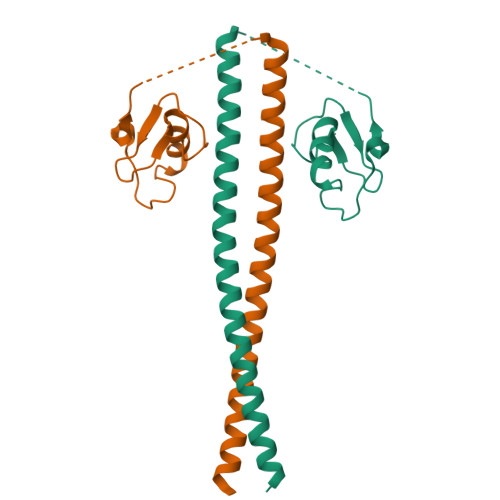
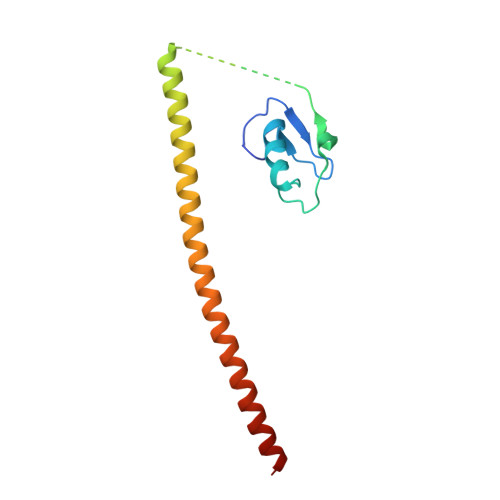
Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
de Moura, T.R., Mozaffari-Jovin, S., Szabo, C.Z.K., Schmitzova, J., Dybkov, O., Cretu, C., Kachala, M., Svergun, D., Urlaub, H., Luhrmann, R., Pena, V.(2018) Mol Cell 69: 979-992.e6
- PubMed: 29547724
- DOI: https://doi.org/10.1016/j.molcel.2018.02.022
- Primary Citation of Related Structures:
5M88, 5M89, 5M8C - PubMed Abstract:
Human nineteen complex (NTC) acts as a multimeric E3 ubiquitin ligase in DNA repair and splicing. The transfer of ubiquitin is mediated by Prp19-a homotetrameric component of NTC whose elongated coiled coils serve as an assembly axis for two other proteins called SPF27 and CDC5L. We find that Prp19 is inactive on its own and have elucidated the structural basis of its autoinhibition by crystallography and mutational analysis. Formation of the NTC core by stepwise assembly of SPF27, CDC5L, and PLRG1 onto the Prp19 tetramer enables ubiquitin ligation. Protein-protein crosslinking of NTC, functional assays in vitro, and assessment of its role in DNA damage response provide mechanistic insight into the organization of the NTC core and the communication between PLRG1 and Prp19 that enables E3 activity. This reveals a unique mode of regulation for a complex E3 ligase and advances understanding of its dynamics in various cellular pathways.
Organizational Affiliation:
Macromolecular Crystallography Group, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.