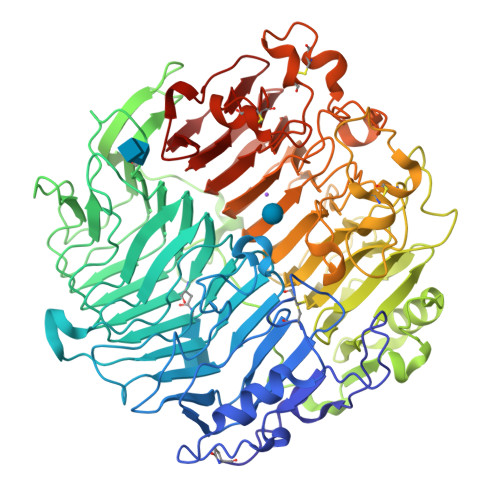
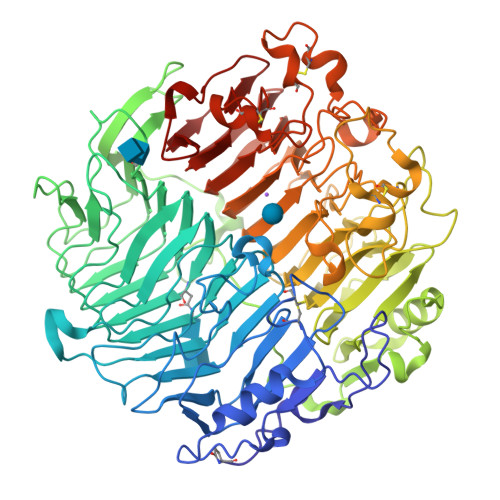
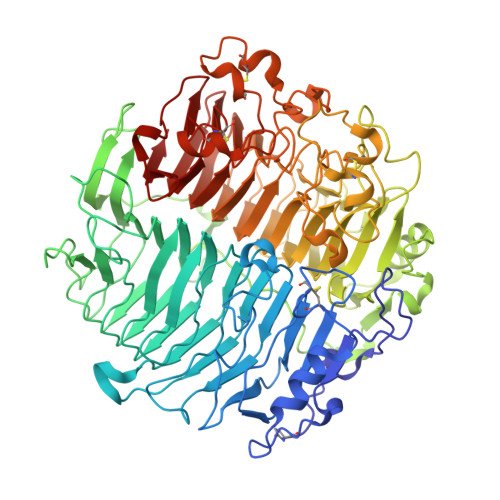
Crystal structure and biological implications of a glycoside hydrolase family 55 beta-1,3-glucanase from Chaetomium thermophilum.
Papageorgiou, A.C., Chen, J., Li, D.(2017) Biochim Biophys Acta 1865: 1030-1038
- PubMed: 28479293
- DOI: https://doi.org/10.1016/j.bbapap.2017.05.002
- Primary Citation of Related Structures:
5M5Z, 5M60 - PubMed Abstract:
Crystal structures of a β-1,3-glucanase from the thermophilic fungus Chaetomium thermophilum were determined at 1.20 and 1.42Å resolution in the free and glucose-bound form, respectively. This is the third structure of a family 55 glycoside hydrolase (GH55) member and the second from a fungus. Based on comparative structural studies and site-directed mutagenesis, Glu654 is proposed as the catalytic acid residue. The substrate binding cleft exhibits restricted access on one side, rendering the enzyme as an exo-β-1,3-glucanase as confirmed also by thin layer chromatography experiments. A lack of stacking interactions was found at the substrate binding cleft, suggesting that interactions at positions -1, +1 and +2 are sufficient to orientate the substrate. A binding pocket was identified that could explain binding of branched laminarin and accumulation of laminaritriose.
Organizational Affiliation:
Turku Centre for Biotechnology, University of Turku and Åbo Akademi University, 20521 Turku, Finland. Electronic address: tassos.papageorgiou@btk.fi.