Structural basis for the shielding function of the dynamic trypanosome variant surface glycoprotein coat.
Bartossek, T., Jones, N.G., Schafer, C., Cvitkovic, M., Glogger, M., Mott, H.R., Kuper, J., Brennich, M., Carrington, M., Smith, A.S., Fenz, S., Kisker, C., Engstler, M.(2017) Nat Microbiol 2: 1523-1532
- PubMed: 28894098
- DOI: https://doi.org/10.1038/s41564-017-0013-6
- Primary Citation of Related Structures:
5LY9, 5M4T - PubMed Abstract:
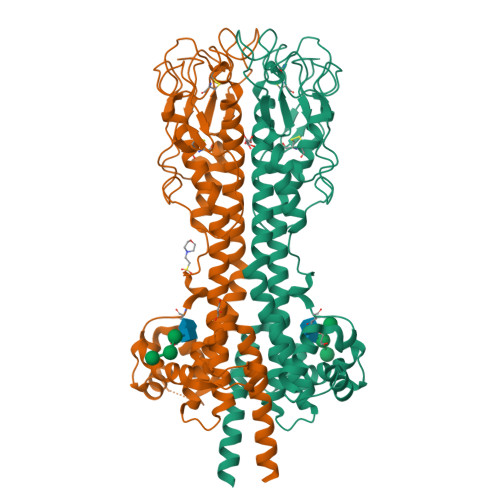
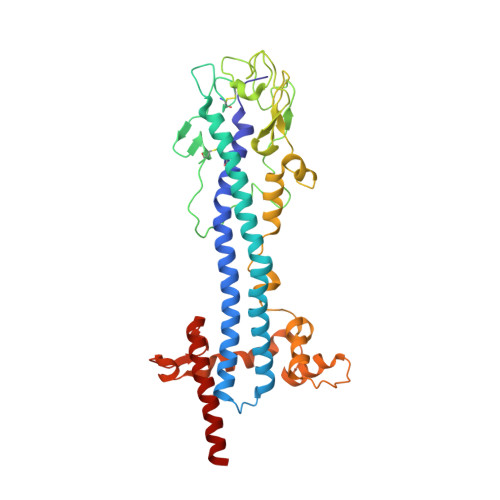
The most prominent defence of the unicellular parasite Trypanosoma brucei against the host immune system is a dense coat that comprises a variant surface glycoprotein (VSG). Despite the importance of the VSG family, no complete structure of a VSG has been reported. Making use of high-resolution structures of individual VSG domains, we employed small-angle X-ray scattering to elucidate the first two complete VSG structures. The resulting models imply that the linker regions confer great flexibility between domains, which suggests that VSGs can adopt two main conformations to respond to obstacles and changes of protein density, while maintaining a protective barrier at all times. Single-molecule diffusion measurements of VSG in supported lipid bilayers substantiate this possibility, as two freely diffusing populations could be detected. This translates into a highly flexible overall topology of the surface VSG coat, which displays both lateral movement in the plane of the membrane and variation in the overall thickness of the coat.
Organizational Affiliation:
Department of Cell and Developmental Biology, Theodor-Boveri-Institute, Biocenter, University of Würzburg, 97074, Würzburg, Germany.