Human NUDT22 Is a UDP-Glucose/Galactose Hydrolase Exhibiting a Unique Structural Fold.
Carter, M., Jemth, A.S., Carreras-Puigvert, J., Herr, P., Martinez Carranza, M., Vallin, K.S.A., Throup, A., Helleday, T., Stenmark, P.(2018) Structure 26: 295-303.e6
- PubMed: 29413322
- DOI: https://doi.org/10.1016/j.str.2018.01.004
- Primary Citation of Related Structures:
5LOU - PubMed Abstract:
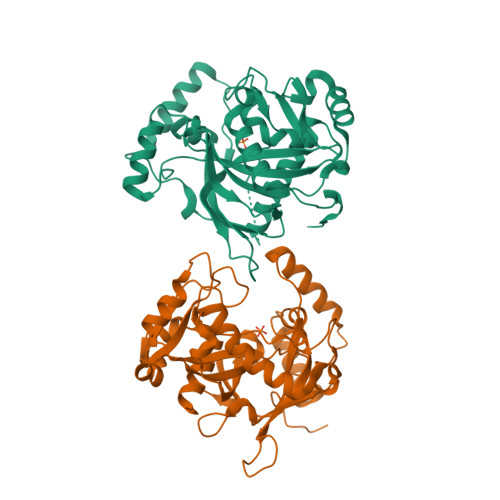
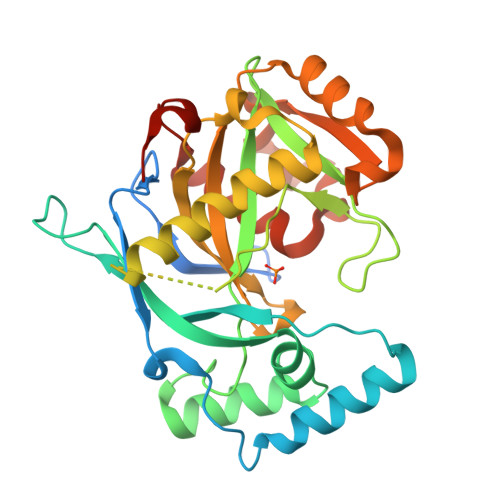
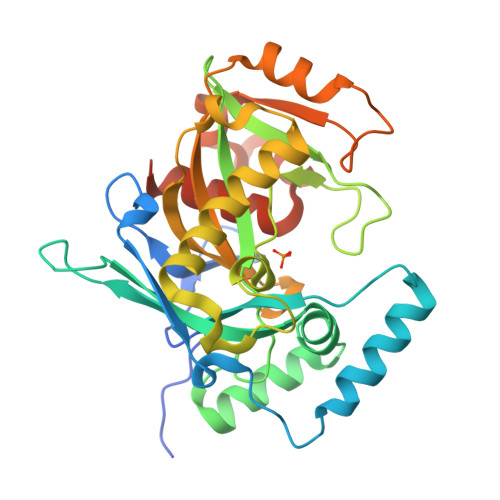
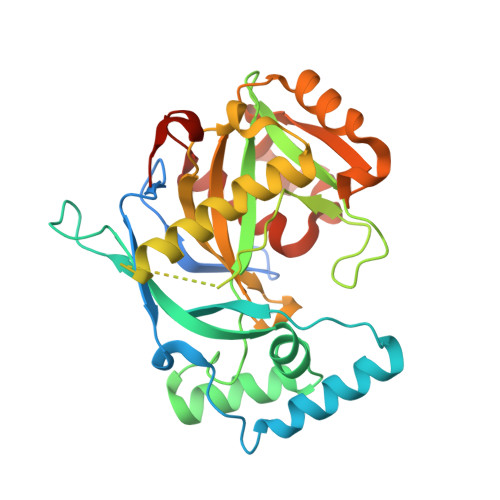
Human NUDT22 belongs to the diverse NUDIX family of proteins, but has, until now, remained uncharacterized. Here we show that human NUDT22 is a Mg 2+ -dependent UDP-glucose and UDP-galactose hydrolase, producing UMP and glucose 1-phosphate or galactose 1-phosphate. We present the structure of human NUDT22 alone and in a complex with the substrate UDP-glucose. These structures reveal a partially conserved NUDIX fold domain preceded by a unique N-terminal domain responsible for UDP moiety binding and recognition. The NUDIX domain of NUDT22 contains a modified NUDIX box identified using structural analysis and confirmed through functional analysis of mutants. Human NUDT22's distinct structure and function as a UDP-carbohydrate hydrolase establish a unique NUDIX protein subfamily.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University, 106 91 Stockholm, Sweden.