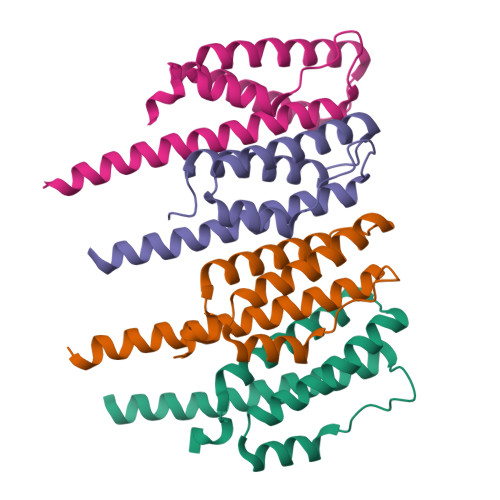
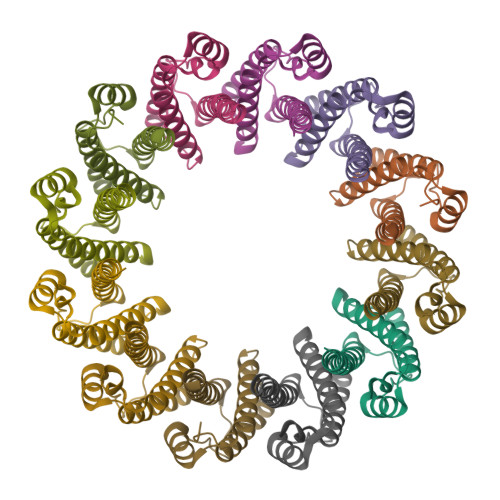
Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Bolten, M., Delley, C.L., Leibundgut, M., Boehringer, D., Ban, N., Weber-Ban, E.(2016) Structure 24: 2138-2151
- PubMed: 27839949
- DOI: https://doi.org/10.1016/j.str.2016.10.008
- Primary Citation of Related Structures:
5LFJ, 5LFP, 5LFQ, 5LZP - PubMed Abstract:
Mycobacterium tuberculosis harbors proteasomes that recruit substrates for degradation through an ubiquitin-like modification pathway. Recently, a non-ATPase activator termed Bpa (bacterial proteasome activator) was shown to support an alternate proteasomal degradation pathway. Here, we present the cryo-electron microscopy (cryo-EM) structure of Bpa in complex with the 20S core particle (CP). For docking into the cryo-EM density, we solved the X-ray structure of Bpa, showing that it forms tight four-helix bundles arranged into a 12-membered ring with a 40 Å wide central pore and the C-terminal helix of each protomer protruding from the ring. The Bpa model was fitted into the cryo-EM map of the Bpa-CP complex, revealing its architecture and striking symmetry mismatch. The Bpa-CP interface was resolved to 3.5 Å, showing the interactions between the C-terminal GQYL motif of Bpa and the proteasome α-rings. This docking mode is related to the one observed for eukaryotic activators with features specific to the bacterial complex.
Organizational Affiliation:
ETH Zurich, Institute of Molecular Biology & Biophysics, 8093 Zurich, Switzerland.