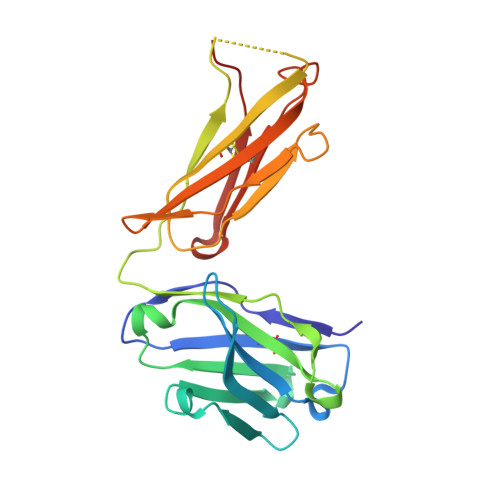
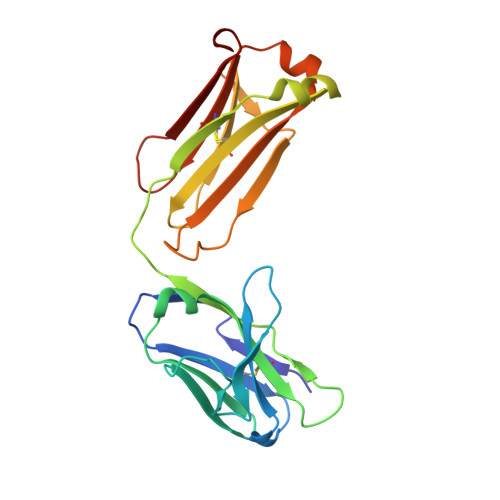
The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Naschberger, A., Furnrohr, B.G., Lenac Rovis, T., Malic, S., Scheffzek, K., Dieplinger, H., Rupp, B.(2016) Acta Crystallogr D Struct Biol 72: 1267-1280
- PubMed: 27917827
- DOI: https://doi.org/10.1107/S205979831601723X
- Primary Citation of Related Structures:
5L7X, 5L88, 5L9D, 5LGH - PubMed Abstract:
The monoclonal antibody N14 is used as a detection antibody in ELISA kits for the human glycoprotein afamin, a member of the albumin family, which has recently gained interest in the capture and stabilization of Wnt signalling proteins, and for its role in metabolic syndrome and papillary thyroid carcinoma. As a rare occurrence, the N14 Fab is N-glycosylated at Asn26L at the onset of the V L 1 antigen-binding loop, with the α-1-6 core fucosylated complex glycan facing out of the L1 complementarity-determining region. The crystal structures of two non-apparent (pseudo) isomorphous crystals of the N14 Fab were analyzed, which differ significantly in the elbow angles, thereby cautioning against the overinterpretation of domain movements upon antigen binding. In addition, the map quality at 1.9 Å resolution was sufficient to crystallographically re-sequence the variable V L and V H domains and to detect discrepancies in the hybridoma-derived sequence. Finally, a conservatively refined parsimonious model is presented and its statistics are compared with those from a less conservatively built model that has been modelled more enthusiastically. Improvements to the PDB validation reports affecting ligands, clashscore and buried surface calculations are suggested.
Organizational Affiliation:
Division of Biological Chemistry, Medical University of Innsbruck, Innrain 80, 6020 Innsbruck, Austria.