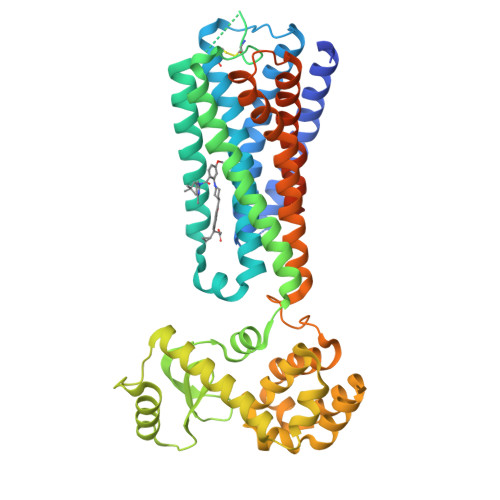
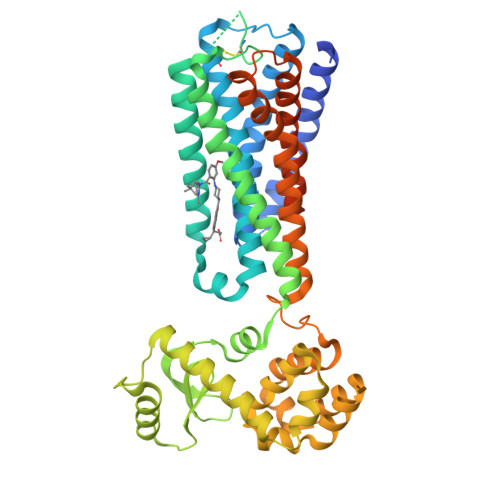
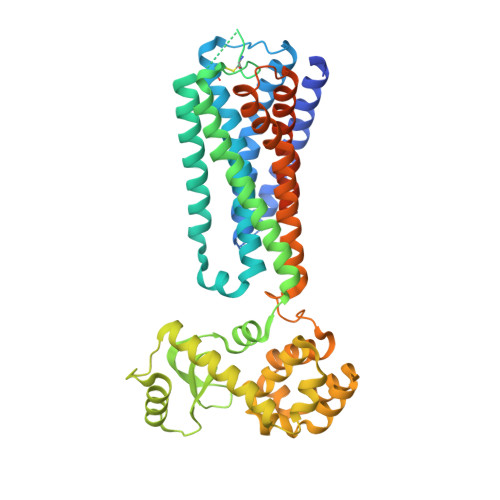
Structural basis for GPR40 allosteric agonism and incretin stimulation.
Ho, J.D., Chau, B., Rodgers, L., Lu, F., Wilbur, K.L., Otto, K.A., Chen, Y., Song, M., Riley, J.P., Yang, H.C., Reynolds, N.A., Kahl, S.D., Lewis, A.P., Groshong, C., Madsen, R.E., Conners, K., Lineswala, J.P., Gheyi, T., Saflor, M.D., Lee, M.R., Benach, J., Baker, K.A., Montrose-Rafizadeh, C., Genin, M.J., Miller, A.R., Hamdouchi, C.(2018) Nat Commun 9: 1645-1645
- PubMed: 29695780
- DOI: https://doi.org/10.1038/s41467-017-01240-w
- Primary Citation of Related Structures:
5KW2 - PubMed Abstract:
Activation of free fatty acid receptor 1 (GPR40) by synthetic partial and full agonists occur via distinct allosteric sites. A crystal structure of GPR40-TAK-875 complex revealed the allosteric site for the partial agonist. Here we report the 2.76-Å crystal structure of human GPR40 in complex with a synthetic full agonist, compound 1, bound to the second allosteric site. Unlike TAK-875, which acts as a Gα q -coupled partial agonist, compound 1 is a dual Gα q and Gα s -coupled full agonist. compound 1 binds in the lipid-rich region of the receptor near intracellular loop 2 (ICL2), in which the stabilization of ICL2 by the ligand is likely the primary mechanism for the enhanced G protein activities. The endogenous free fatty acid (FFA), γ-linolenic acid, can be computationally modeled in this site. Both γ-linolenic acid and compound 1 exhibit positive cooperativity with TAK-875, suggesting that this site could also serve as a FFA binding site.
Organizational Affiliation:
Lilly Biotechnology Center San Diego, 10290 Campus Point Drive, San Diego, CA, 92121, USA. ho_joseph_d@lilly.com.