Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Salinger, A.J., Thoden, J.B., Holden, H.M.(2016) Biochemistry 55: 4509-4518
- PubMed: 27404806
- DOI: https://doi.org/10.1021/acs.biochem.6b00602
- Primary Citation of Related Structures:
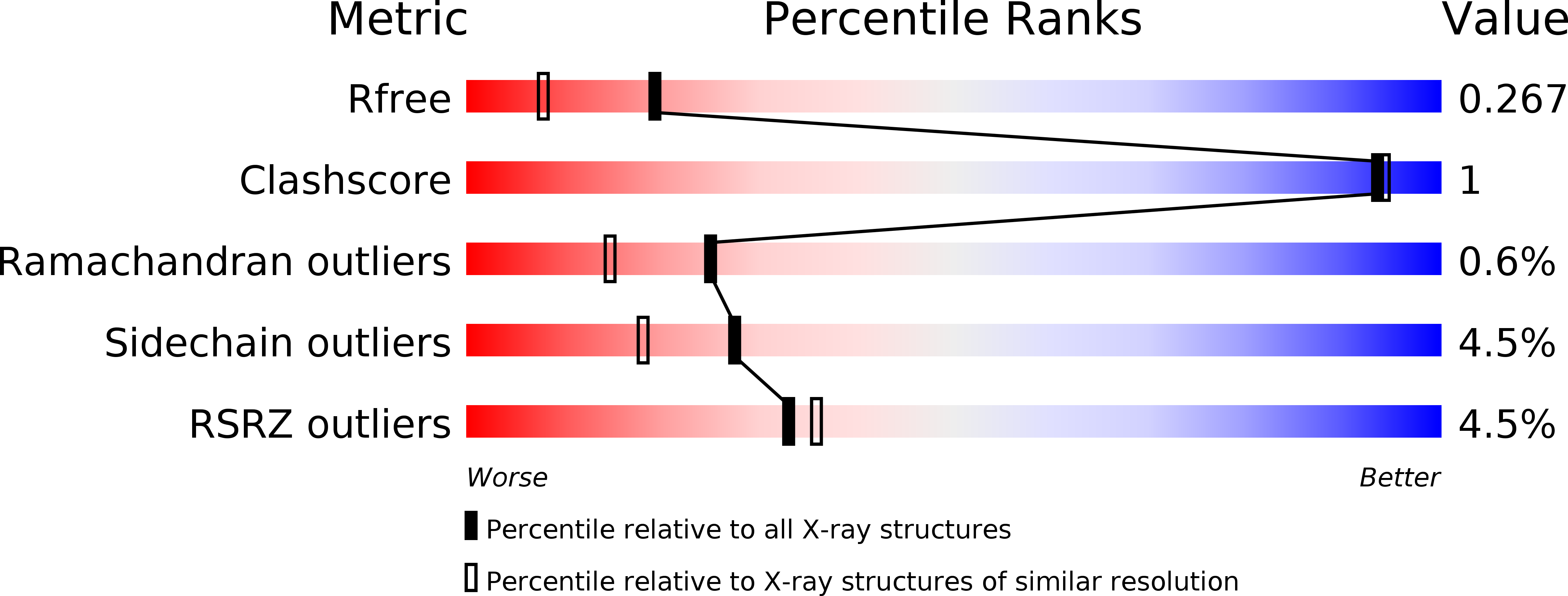
5KTA, 5KTC, 5KTD - PubMed Abstract:
Enzymes belonging to the GNAT superfamily are widely distributed in nature where they play key roles in the transfer of acyl groups from acyl-CoAs to primary amine acceptors. The amine acceptors run the gamut from histones to aminoglycoside antibiotics to small molecules such as serotonin. Whereas those family members that function on histones have been extensively studied, the GNAT enzymes that employ nucleotide-linked sugars as their substrates have not been well characterized. Indeed, though the structures of two of these "amino sugar" GNAT enzymes have been determined within the past 10 years, details concerning their active site architectures have been limited because of a lack of bound nucleotide-linked sugar substrates. Here we describe a combined structural and biochemical analysis of FdhC from Acinetobacter nosocomialis O2. On the basis of bioinformatics, it was postulated that FdhC catalyzes the transfer of a 3-hydroxybutanoyl group from 3-hydroxylbutanoyl-CoA to dTDP-3-amino-3,6-dideoxy-d-galactose, to yield an unusual sugar that is ultimately incorporated into the surface polysaccharides of the bacterium. We present data confirming this activity. In addition, the structures of two ternary complexes of FdhC, in the presence of CoA and either 3-hydroxybutanoylamino-3,6-dideoxy-d-galactose or 3-hydroxybutanoylamino-3,6-dideoxy-d-glucose, were solved by X-ray crystallographic analyses to high resolution. Kinetic parameters were determined, and activity assays demonstrated that FdhC can also utilize acetyl-CoA, 3-methylcrotonyl-CoA, or hexanoyl-CoA as acyl donors, albeit at reduced rates. Site-directed mutagenesis experiments were conducted to probe the catalytic mechanism of FdhC. Taken together, the data presented herein provide significantly new molecular insight into those GNAT superfamily members that function on nucleotide-linked amino sugars.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin , Madison, Wisconsin 53706, United States.