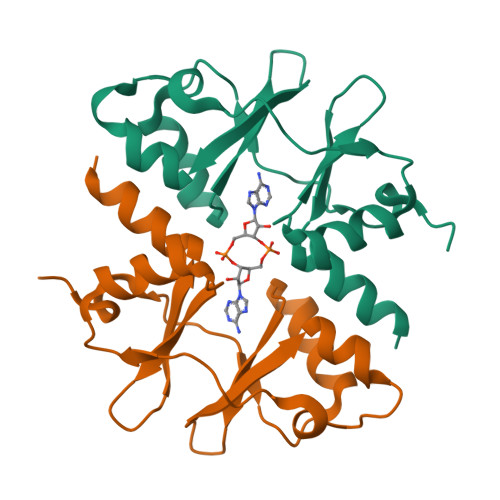
Cyclic di-AMP targets the cystathionine beta-synthase domain of the osmolyte transporter OpuC.
Huynh, T.N., Choi, P.H., Sureka, K., Ledvina, H.E., Campillo, J., Tong, L., Woodward, J.J.(2016) Mol Microbiol 102: 233-243
- PubMed: 27378384
- DOI: https://doi.org/10.1111/mmi.13456
- Primary Citation of Related Structures:
5KS7 - PubMed Abstract:
Cellular turgor is of fundamental importance to bacterial growth and survival. Changes in external osmolarity as a consequence of fluctuating environmental conditions and colonization of diverse environments can significantly impact cytoplasmic water content, resulting in cellular lysis or plasmolysis. To ensure maintenance of appropriate cellular turgor, bacteria import ions and small organic osmolytes, deemed compatible solutes, to equilibrate cytoplasmic osmolarity with the extracellular environment. Here, we show that elevated levels of c-di-AMP, a ubiquitous second messenger among bacteria, result in significant susceptibility to elevated osmotic stress in the bacterial pathogen Listeria monocytogenes. We found that levels of import of the compatible solute carnitine show an inverse correlation with intracellular c-di-AMP content and that c-di-AMP directly binds to the CBS domain of the ATPase subunit of the carnitine importer OpuC. Biochemical and structural studies identify conserved residues required for this interaction and transport activity in bacterial cells. Overall, these studies reveal a role for c-di-AMP mediated regulation of compatible solute import and provide new insight into the molecular mechanisms by which this essential second messenger impacts bacterial physiology and adaptation to changing environmental conditions.
Organizational Affiliation:
Department of Microbiology, University of Washington, Seattle, WA, 98195, USA.