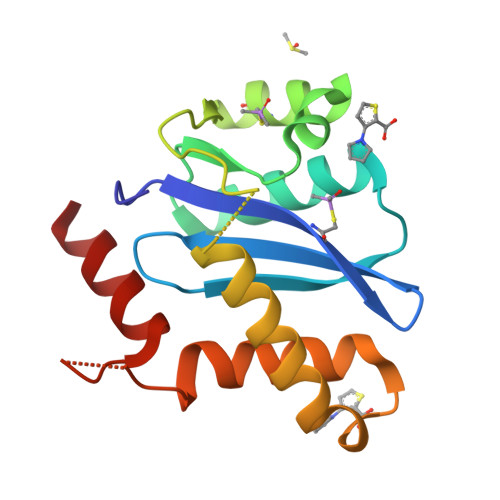
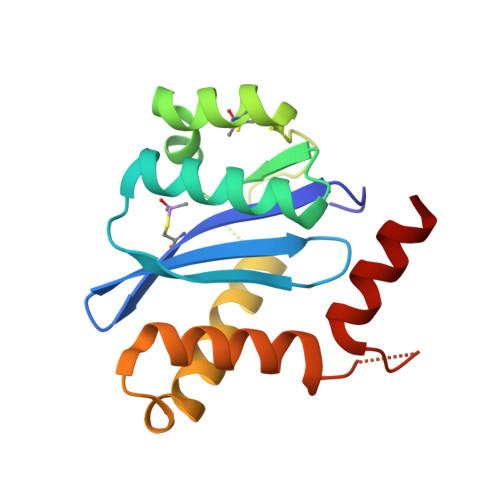
A New Class of Allosteric HIV-1 Integrase Inhibitors Identified by Crystallographic Fragment Screening of the Catalytic Core Domain.
Patel, D., Antwi, J., Koneru, P.C., Serrao, E., Forli, S., Kessl, J.J., Feng, L., Deng, N., Levy, R.M., Fuchs, J.R., Olson, A.J., Engelman, A.N., Bauman, J.D., Kvaratskhelia, M., Arnold, E.(2016) J Biol Chem 291: 23569-23577
- PubMed: 27645997
- DOI: https://doi.org/10.1074/jbc.M116.753384
- Primary Citation of Related Structures:
5KRS, 5KRT - PubMed Abstract:
HIV-1 integrase (IN) is essential for virus replication and represents an important multifunctional therapeutic target. Recently discovered quinoline-based allosteric IN inhibitors (ALLINIs) potently impair HIV-1 replication and are currently in clinical trials. ALLINIs exhibit a multimodal mechanism of action by inducing aberrant IN multimerization during virion morphogenesis and by competing with IN for binding to its cognate cellular cofactor LEDGF/p75 during early steps of HIV-1 infection. However, quinoline-based ALLINIs impose a low genetic barrier for the evolution of resistant phenotypes, which highlights a need for discovery of second-generation inhibitors. Using crystallographic screening of a library of 971 fragments against the HIV-1 IN catalytic core domain (CCD) followed by a fragment expansion approach, we have identified thiophenecarboxylic acid derivatives that bind at the CCD-CCD dimer interface at the principal lens epithelium-derived growth factor (LEDGF)/p75 binding pocket. The most active derivative (5) inhibited LEDGF/p75-dependent HIV-1 IN activity in vitro with an IC 50 of 72 μm and impaired HIV-1 infection of T cells at an EC 50 of 36 μm The identified lead compound, with a relatively small molecular weight (221 Da), provides an optimal building block for developing a new class of inhibitors. Furthermore, although structurally distinct thiophenecarboxylic acid derivatives target a similar pocket at the IN dimer interface as the quinoline-based ALLINIs, the lead compound, 5, inhibited IN mutants that confer resistance to quinoline-based compounds. Collectively, our findings provide a plausible path for structure-based development of second-generation ALLINIs.
Organizational Affiliation:
From the Center for Advanced Biotechnology and Medicine, Rutgers University, Piscataway, New Jersey 08854.