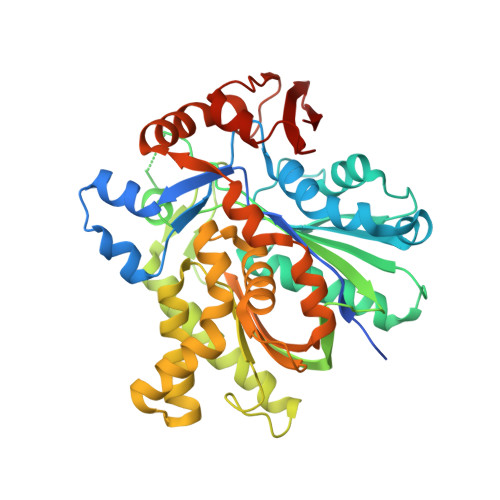
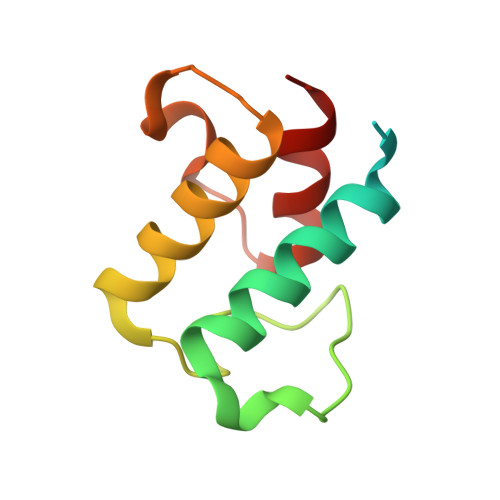
Anatomy of the beta-branching enzyme of polyketide biosynthesis and its interaction with an acyl-ACP substrate.
Maloney, F.P., Gerwick, L., Gerwick, W.H., Sherman, D.H., Smith, J.L.(2016) Proc Natl Acad Sci U S A 113: 10316-10321
- PubMed: 27573844
- DOI: https://doi.org/10.1073/pnas.1607210113
- Primary Citation of Related Structures:
5KP5, 5KP6, 5KP7, 5KP8 - PubMed Abstract:
Alkyl branching at the β position of a polyketide intermediate is an important variation on canonical polyketide natural product biosynthesis. The branching enzyme, 3-hydroxy-3-methylglutaryl synthase (HMGS), catalyzes the aldol addition of an acyl donor to a β-keto-polyketide intermediate acceptor. HMGS is highly selective for two specialized acyl carrier proteins (ACPs) that deliver the donor and acceptor substrates. The HMGS from the curacin A biosynthetic pathway (CurD) was examined to establish the basis for ACP selectivity. The donor ACP (CurB) had high affinity for the enzyme (Kd = 0.5 μM) and could not be substituted by the acceptor ACP. High-resolution crystal structures of HMGS alone and in complex with its donor ACP reveal a tight interaction that depends on exquisite surface shape and charge complementarity between the proteins. Selectivity is explained by HMGS binding to an unusual surface cleft on the donor ACP, in a manner that would exclude the acceptor ACP. Within the active site, HMGS discriminates between pre- and postreaction states of the donor ACP. The free phosphopantetheine (Ppant) cofactor of ACP occupies a conserved pocket that excludes the acetyl-Ppant substrate. In comparison with HMG-CoA (CoA) synthase, the homologous enzyme from primary metabolism, HMGS has several differences at the active site entrance, including a flexible-loop insertion, which may account for the specificity of one enzyme for substrates delivered by ACP and the other by CoA.
Organizational Affiliation:
Life Sciences Institute, University of Michigan, Ann Arbor, MI 48109; Chemical Biology Doctoral Program, University of Michigan, Ann Arbor, MI 48109;