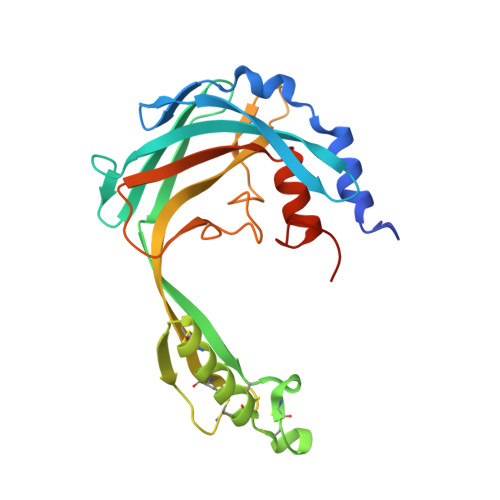
Structural characterization reveals a novel bilobed architecture for the ectodomains of insect stage expressed Trypanosoma brucei PSSA-2 and Trypanosoma congolense ISA.
Ramaswamy, R., Goomeshi Nobary, S., Eyford, B.A., Pearson, T.W., Boulanger, M.J.(2016) Protein Sci 25: 2297-2302
- PubMed: 27671214
- DOI: https://doi.org/10.1002/pro.3053
- Primary Citation of Related Structures:
5KLH, 5KMX - PubMed Abstract:
African trypanosomiasis, caused by parasites of the genus Trypanosoma, is a complex of devastating vector-borne diseases of humans and livestock in sub-Saharan Africa. Central to the pathogenesis of African trypanosomes is their transmission by the arthropod vector, Glossina spp. (tsetse fly). Intriguingly, the efficiency of parasite transmission through the vector is reduced following depletion of Trypanosoma brucei Procyclic-Specific Surface Antigen-2 (TbPSSA-2). To investigate the underlying molecular mechanism of TbPSSA-2, we determined the crystal structures of its ectodomain and that of its homolog T. congolense Insect Stage Antigen (TcISA) to resolutions of 1.65 Å and 2.45 Å, respectively using single wavelength anomalous dispersion. Both proteins adopt a novel bilobed architecture with the individual lobes displaying rotational flexibility around the central tether that suggest a potential mechanism for coordinating a binding partner. In support of this hypothesis, electron density consistent with a bound peptide was observed in the inter-lob cleft of a TcISA monomer. These first reported structures of insect stage transmembrane proteins expressed by African trypanosomes provide potentially valuable insight into the interface between parasite and tsetse vector.
Organizational Affiliation:
Department of Biochemistry and Microbiology, University of Victoria, Victoria, BC, Canada, V8W 3P6.